Translate this page into:
Exploring the therapeutic efficacy: Unveiling the active compounds of Huashi Baidu granules against COVID-19
⁎Corresponding author at: Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China. yyzhou@icmm.ac.cn (Yanyan Zhou),
⁎⁎Corresponding author at: Guang’anmen Hospital, China Academy of Chinese Medical Sciences, Beijing 100053, China limin-72114@163.com (Min Li)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
Huashi Baidu granule (HSBD), an approved herbal formula for treating COVID-19, demonstrates safety and efficacy. Despite its market approval, the detailed methodology and identification of its active components remain unexplored, leaving its bioactive constituents and action mechanisms unclear.
Methods
This study investigated the potential mechanisms of HSBD’s active ingredient in treating COVID-19. Our approach integrated various techniques, including the UHPLC-QqQ-MS/MS method, analysis of the GEO database, network pharmacology, surface plasmon resonance, molecular docking and molecular dynamics simulations, to formulate a comprehensive research strategy.
Results
The UHPLC-QqQ-MS/MS method employed for HSBD analysis proved stable, reliable, and reproducible. We identified 25 principal components in HSBD, with 7 compounds detected in plasma, namely Pogostone, P-Hydroxybenzoic acid, Paeoniflorin, Rhein, Emodin, Ephedrine hydrochloride, and Pseudoephedrine hydrochloride. Protein-Protein Interaction (PPI) network analysis identified MMP9 as a pivotal target. Surface plasmon resonance analysis revealed that Paeoniflorin and Rhein exert their antiviral effects by interacting with RBD and ACE2. In contrast, Emodin’s antiviral mechanism predominantly involves binding to MMP9. Molecular docking results indicated strong binding affinities of Rhein and Paeoniflorin to the hACE2 protein, and high binding affinities of Emodin to the MMP9 protein, all of which were corroborated by molecular dynamics simulations.
Conclusion
We investigated the methodology and identified the active components of HSBD, focusing on those absorbed into the plasma, to elucidate the effective material basis of HSBD in the treatment of COVID-19, our research offered insightful exploration into its mechanisms of action against COVID-19.
Keywords
Huashi Baidu granule (HSBD)
COVID-19
Molecular docking
Surface plasmon resonance (SPR)
Protein-Protein Interaction (PPI) network
Data availability
The data that support the findings of this study are available from the corresponding author on reasonable request.
1 Introduction
COVID-19, caused by the SARS-CoV-2 virus, has been a global health crisis since December 2019(Zhou et al., 2020, Markov et al., 2023), with over 760 million cases and 6.9 million fatalities as reported by the World Health Organization. In this context, Huashi Baidu granule (HSBD), also known as Q-14, comprises 14 Chinese materia medica and has significantly contributed to COVID-19 treatment in China(Yu et al., 2023, Chen et al., 2023). Following its approval by the State Drug Administration on March 2, 2021. HSBD has demonstrated considerable efficacy in reducing clinical symptoms, improving inflammatory markers and oxygenation levels, enhancing viral clearance, and promoting lung recovery, thus confirming its safety and effectiveness for severe COVID-19 cases(Wang et al., 2021b, Xiong et al., 2022). Previous studies have shown that the bioactive compounds in HSBD exhibit both antiviral and anti-inflammatory effects against COVID-19(Xu et al., 2023). Additionally, HSBD has been proven to inhibit cytokine storms and reduce inflammatory symptoms via the TLR4/NF-κB and PI3K/Akt pathways(Zhang et al., 2023). However, despite its clinical success and the findings of previous studies, the specifics of HSBD's active components and their mechanisms remain inadequately explored.
We have used the the Ultra-High-Performance Liquid Chromatography-Triple Quadrupole Mass Spectrometry (UHPLC-QqQ-MS/MS) technique to profile the active compounds in HSBD, especially those compounds that were absorbed into plasma. On the basis of this, our study adopts an integrated research approach, combining the GEO database, network pharmacology, surface plasmon resonance, molecular docking and molecular dynamics simulation, to elucidate the underlying mechanisms of the active ingredients of HSBD in combating COVID-19, the study's flow chart is displayed in Figure S1.
2 Materials and methods
2.1 Materials and chemicals
Ephedrine Hydrochloride, Pseudoephedrine Hydrochloride, Amygdalin, Glycyrrhizin, Paeoniflorin, and Astragaloside were acquired from the China Academy of Food and Drug Testing and Research. Additional compounds such as Isorhamnetin-3-O-beta-D-Glucoside, Hyperoside, Sinapic acid, Pogostone, Pachymic acid, Magnolol, Calycosin-7-O-beta-D-glucoside, Honokiol, P-Hydroxybenzoic acid, Quercetin-3-O-beta-D-glucose-7-O-beta-D-gentiobioside, Gentianic acid, Glycyrrhetinic acid, Atractylon, and Hederagenin were sourced from Beijing Saibaike Technology Co. AloeEmodin, Rhein, and Physcion were procured from Chengdu Ruifenshi Biotechnology Co. Meanwhile, Chrysophanic acid and Emodin were obtained from Shanghai Yuanye Biotechnology Co., and CinobufaginIS was purchased from the China National Institute for the Control of Pharmaceutical and Biological Products. Glibenclamide IS was acquired from Shanghai Aladdin Biochemical Technology Co.
HSBD were supplied by Guangdong E Fang Pharmaceutical Co. High-quality reagents, including methanol, formic acid, acetonitrile from Thermo Fisher Scientific (China) Co. Ltd, distilled water from Watson's, and other analytically pure reagents, were utilized to ensure the integrity of the research.
2.2 Chromatographic conditions
Chromatographic conditions were specified using an ACQUITY UHPLC HSS T3 column (2.1 mm × 100 mm, 1.8 µm). The mobile phase consisted of acetonitrile–water with 0.1 % formic acid. The gradient elution profile was as follows: from 0 to 3 min, 1 % acetonitrile; 3 to 5 min, 1 % to 30 % acetonitrile; 5 to 7 min, 30 % to 35 % acetonitrile; 7 to 12 min, 35 % to 95 % acetonitrile; 12 to 13 min, maintained at 95 % acetonitrile; and 13 to 16 min, 95 % back to 1 % acetonitrile. The flow rate was set at 0.25 mL/min, with a column temperature of 30 °C and an injection volume of 2 µL.
For mass spectrometry, an electrospray ionization ion source (ESI) was used in multiple reaction monitoring (MRM) mode with detection capabilities in both positive and negative ion modes. The spray voltage (ion spray) was set at 4500 V, the ion source temperature at 550 °C, and the collision gas (CAD) flow rate at 9 psi (approximately 6.895 kPa). The curtain gas (CUR) was set at 20 psi, and the nebulized gas (N2) at 14 psi. Specific conditions for ion pair detection are detailed in Table S1.
2.3 Standard solution preparation
Accurately measure the required quantities of each standard and dissolve them in chromatographic grade methanol to prepare individual standard stock solutions. Subsequently, combine and dilute these stock solutions sequentially to obtain a mixed standard solution. For the internal standard preparation, progressively dilute the glibenclamide and huazenin toxic base stock solutions to final concentrations of 1.5 µg/mL and 2.5 µg/mL, respectively. Store all prepared solutions at −20 °C until analysis.
2.4 Solution preparation
2.4.1 Test material
Dissolve 0.5 g of the sample powder in 25 mL of 70 % methanol, followed by ultrasonic extraction for 30 min and subsequent filtration. Aliquot 980 µL of the resulting supernatant and enrich it with 10 µL of glibenclamide internal standard solution and 10 µL of huazenin toxic base internal standard solution. Thoroughly vortex the mixture to ensure homogeneity, then transfer it into a liquid phase vial to facilitate quantitative analysis.
2.4.2 Plasma sample preparation
Twelve SPF male Wistar rats, weighing between 180 and 220 g, were approved by the Animal Ethics Committee of Guang'anmen Hospital, China Academy of Traditional Chinese Medicine (IACUC-GANH-2020–008). The rats were housed in the Medical Experiment Center of the China Academy of Traditional Chinese Medicine, in accordance with the Regulations for the Administration of Laboratory Animals of the People's Republic of China. The dose of HSBD used in the rats was converted from the clinical dose using the following formula: HSBD clinical equivalent dose = 20 g × 6.3 / 70 kg = 1.8 g/kg. Humans were given 20 g of HSBD daily, with an average body weight of 70 kg, resulting in a human-to-rat dose ratio of 6.3. The twelve rats were randomly assigned to two groups, with free access to water and food. After a 3-day acclimatization period, the blank group received pure water by gavage, while the HSBD group received 1.8 g/kg HSBD by gavage daily for 7 days. Blood was collected under anesthesia 2 h after the last dose. Blood was drawn from the aorta into a negative pressure tube containing sodium heparin, centrifuged, and the plasma was collected and stored in a −80℃ freezer.
Transfer 200 μL of plasma into a 1.5 mL centrifuge tube, then add 600 μL of acetonitrile. Vortex mix the contents for 3 min, followed by centrifugation at 12,000 rpm for 15 min at 4 ℃. Carefully collect the supernatant and place it into a liquid phase vial for subsequent analysis.
2.5 Differential gene acquisition in COVID-19 severe patients
The GSE164805 dataset, sourced from the GEO database (https://www.ncbi.nlm.nih.gov/geo/), comprises 15 samples, including five patients from each of the following categories: healthy, mild, and severe. To identify differential genes in severe COVID-19 patients, 5 healthy and 5 severe samples were selected for analysis. Using the GEO2R online tool, we screened differentially expressed genes (DEGs) between healthy and severe COVID-19 patients. Differential analysis was conducted with the R package “limma,” setting thresholds for differential genes at |logFC| > 2 and p-value < 0.05. Genes meeting these criteria were considered to exhibit significant differential expression and were identified as potential drug targets in severe COVID-19 patients.
2.6 Target construction components absorbed into the plasma of HSBD
The Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) was queried for seven components: Pogostone, P-Hydroxybenzoic acid, Paeoniflorin, Rhein, Emodin, Ephedrine Hydrochloride, and Pseudoephedrine Hydrochloride. These components were screened to identify those with an oral bioavailability (OB) of at least 30 % and a drug-likeness (DL) value of 0.18 or higher. Subsequently, information on the targets associated with these components was extracted. The protein names of these targets were then converted to gene names utilizing the UniProt database.
2.7 Construction of protein–protein interaction networks and identification of hub genes
The STRING platform was utilized to analyze the protein–protein interaction (PPI) network for the intersecting targets of the seven components absorbed into the plasma from HSBD and associated with severe COVID-19. An interaction threshold of 0.4 was established to exclude isolated nodes, with other settings at default values. The resulting data were then imported into Cytoscape 3.8.2 for comprehensive network analysis, where nodes represent distinct targets, and edges illustrate the interactions between these targets.
2.8 Gene ontology and kyoto gene encyclopedia and genome enrichment analysis
The intersecting targets of the seven HSBD components and severe COVID-19 were inputted into the DAVID database, a web-based functional annotation tool. Settings were adjusted to “official gene symbol” for the select identifier and “genelist” for both list type and species, specifying “Homo sapiens” as the species. This setup facilitated the Gene Ontology (GO) enrichment analysis across three categories: biological process, cellular component, and molecular function. Additionally, a KEGG pathway enrichment analysis was conducted. The results of these analyses were subsequently visualized using bubble mapping on the hiplot platform.
2.9 Surface plasmon resonance
The binding affinities of Paeoniflorin, Emodin, and Rhein to hACE2, SARS-CoV-2 S-RBD, and MMP9 were assessed using the Biacore 8 K+instrument (Cytiva, America). Proteins hACE2, SARS-CoV-2 S-RBD, and MMP9, all sourced from Sino Biological, Beijing, China, were immobilized on a CM5 sensor chip. The immobilization levels for these proteins were approximately 10,000 response units (RU). A series of compound concentrations, each containing 5 % DMSO, were sequentially introduced into the channel to ascertain binding affinity. The dissociation constants (KD) for the compounds were calculated by fitting the data to a steady-state affinity model using the Biacore 8 K+Evaluation Software.
In the competitive inhibition assays, hACE2 or S-RBD proteins were immobilized on the CM5 sensor chip via amine-coupling, with immobilization levels around 3,000 RU. Paeoniflorin and Rhein were initially injected for pre-incubation with hACE2 or S-RBD proteins, respectively. Subsequent injections of 5 nM S-RBD or 30 nM hACE2 were administered to evaluate their interaction with hACE2 or S-RBD in the presence of Paeoniflorin or Rhein. The efficacy of inhibition was determined by comparing response units with and without the active compounds.
2.10 Enzymatic assay
The enzymatic activities of SARS-CoV-2 3CLpro, PLpro, and TMPRSS2 were evaluated using the respective peptides Thr-Ser-Ala-Val-Leu-Gln-pNA, Z-Arg-Leu-Arg-Gly-Gly-AMC (supplied by Bachem Bioscience), and Boc-Gln-Ala-Arg-AMC (provided by CUSABIO). The reaction buffer for 3CLpro contained 40 mM PBS (pH 7.3), 100 mM NaCl, 1 mM EDTA, and 0.1 % Triton X-100. For the PLpro assay, the buffer was composed of 50 mM HEPES (pH 7.4), 100 mM NaCl, and 0.01 % Triton X-100 (v/v). The buffer for the TMPRSS2 assay included 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 0.01 % Triton X-100 (v/v). Measurements of absorbance or fluorescence were conducted using a Biotek Synergy Neo2 instrument.
2.11 Molecular docking
The molecular structure files for MMP9 (PDB codes: 8D35, 2OVX), human angiotensin-converting enzyme 2 (hACE2) (PDB code: 3D0G), and the SARS-CoV-2 spike receptor-binding domain (RBD) (PDB code: 7CH5) were retrieved from the Protein Data Bank (PDB) and AlphaFold databases. Subsequent preprocessing of these target proteins involved the removal of water molecules and proto-ligands, utilizing PyMOL software version 2.3.0. Additionally, the small molecular structures of Paeoniflorin (Compound CID: 442534), Rhein (Compound CID: 10168), and Emodin (Compound CID: 3220) were acquired from the PubChem database. Following the download, these small molecules were optimized for their conformations through molecular mechanics calculations using Chem3D software (version 2020), resulting in structures with minimized energy.
The prepared protein molecules were then hydrogenated using AutoDock Tools version 1.5.6. Similarly, the small molecules, post-optimization, were hydrogenated and their torsional bonds were identified. Docking simulations between the target proteins and the small molecules were performed using AutoDock Vina software version 1.2.0. The docking employed a Lamarckian Genetic Algorithm (LGA) in a semi-flexible docking mode, with the exhaustiveness parameter set to 128, to predict the interaction conformations.
2.12 Molecular dynamics simulation
Molecular dynamics simulations were conducted using Gromacs software version 2022.3. The small molecules underwent preprocessing with AmberTools22 to incorporate the GAFF force field, while Gaussian 16 W facilitated their hydrogenation and the calculation of RESP potentials. Simulation conditions were set at a constant temperature of 300 K and atmospheric pressure (1 bar), employing the Amber99sb-ildn force field and the Tip3p water model as the solvent. The system's total charge was neutralized by adding an appropriate quantity of Na + ions.
Initially, the molecular dynamics simulation system underwent energy minimization using the steepest descent method. This was followed by equilibration phases: an isothermal-isochoric (NVT) and an isothermal-isobaric (NPT) equilibration, conducted over 100,000 steps with a coupling constant of 0.1 ps and a duration of 100 ps, respectively. Subsequently, an unrestrained molecular dynamics simulation was executed over 5,000,000 steps with a time step of 2 fs, totaling a simulation time of 100 ns.
Upon completing the simulations, trajectory analysis was performed using the software’s integrated tools to calculate the root mean square deviation (RMSD), root mean square fluctuation (RMSF), and the radius of gyration for each amino acid within the trajectories.
3 Results
3.1 Methodological examination
3.1.1 Linear relationship
The internal standard method was employed to quantify 25 compounds in HSBD samples. Following the protocol delineated in section 2.3, each standard solution was serially diluted. For analysis, 980 µL of each diluted solution was combined with 10 µL of glibenclamide and 10 µL of huazenin toxicity internal standard solutions. The peak areas were measured under conditions specified in section 2.2. The ratio of peak area of each compound to that of the internal standard (Yi/Ys) was calculated to conduct linear regression, facilitating the construction of standard curves and derivation of regression equations and correlation coefficients, which are tabulated in Table S2.
Upon plotting these standard curves, the 25 compounds exhibited strong linear relationships, with correlation coefficients (r values) exceeding 0.99, indicating a high degree of linearity. These findings underscore the effectiveness of the method in quantifying the compounds, as reflected by the substantial correlation coefficients.
3.1.2 Precision, stability and reproducibility study
Pipette 960 μL of the standard mixed solution, then add 10 µL of glibenclamide and 10 µL of huazufenin as internal standard solutions. Following this, perform continuous injections six times within one day under specified conditions (Section 2.2), and subsequently record the peak areas for the 25 active components alongside the internal standards. Utilize the standard curve to determine the concentrations, revealing that the relative standard deviation (RSD) ranges from 0.94 % to 14.24 %. This range suggests satisfactory precision of the instrument.
For reproducibility assessment, precisely weigh six samples of HSBD, prepare the test solutions according to Section 2.4, and measure the peak areas following Section 2.2. The calculation of the RSD for the 25 active ingredients indicates a range of 1.49 % to 9.04 %, confirming the method's reproducibility.
To prepare the high-sensitivity bio-detection test solution, adhere to the guidelines in Section 2.4. Conduct measurements at 0, 2, 4, 8, 12, and 24 h as per Section 2.2, recording the peak areas of the 25 active components and internal standards. Calculation of the concentrations using the standard curve indicates an RSD range from 1.08 % to 5.97 %, demonstrating that the sample maintains stability over a 24-hour period (refer to Table S3).
3.1.3 Sample recovery test
Accurately weigh six samples from the same batch of High Sensitivity Bio-Detection, each weighing 0.5 g, and add a control solution equivalent to the sample content. Prepare the test solution as per the protocol outlined in Section 2.4.1. Following the chromatographic conditions specified in Section 2.2, the average recovery rates of the samples were determined to be between 93.22 % and 98.29 %. The relative standard deviations (RSDs) ranged from 0.8 % to 3.8 %. These findings suggest that the method is both accurate and reliable (refer to Table S4).
3.1.4 Determination of sample content components
The UHPLC-QqQ-MS/MS technique was employed to quantify 25 primary active compounds in the HSBD granule sample, utilizing Multiple Reaction Monitoring (MRM) mode. A 0.5 g sample of HSBD was precisely weighed, and the test solution was prepared following the protocol in Section 2.4.1. Chromatographic conditions were set according to Section 2.2, and the content of each compound was calculated using the corresponding standard curve. The findings are presented in Table S5.
Analysis revealed that seven constituents, including Patchouli ketone, P-Hydroxybenzoic acid (grasshopper ketone), Paeoniflorin, Rhodacanthic acid, Rhubarbine, Ephedrine hydrochloride, and Pseudoephedrine hydrochloride, were detectable in the plasma (Table S6). Fig. 1A displays the MRM profiles for these 25 compounds and the internal standard in the Resolving Dampness and Defeating Toxin Granules.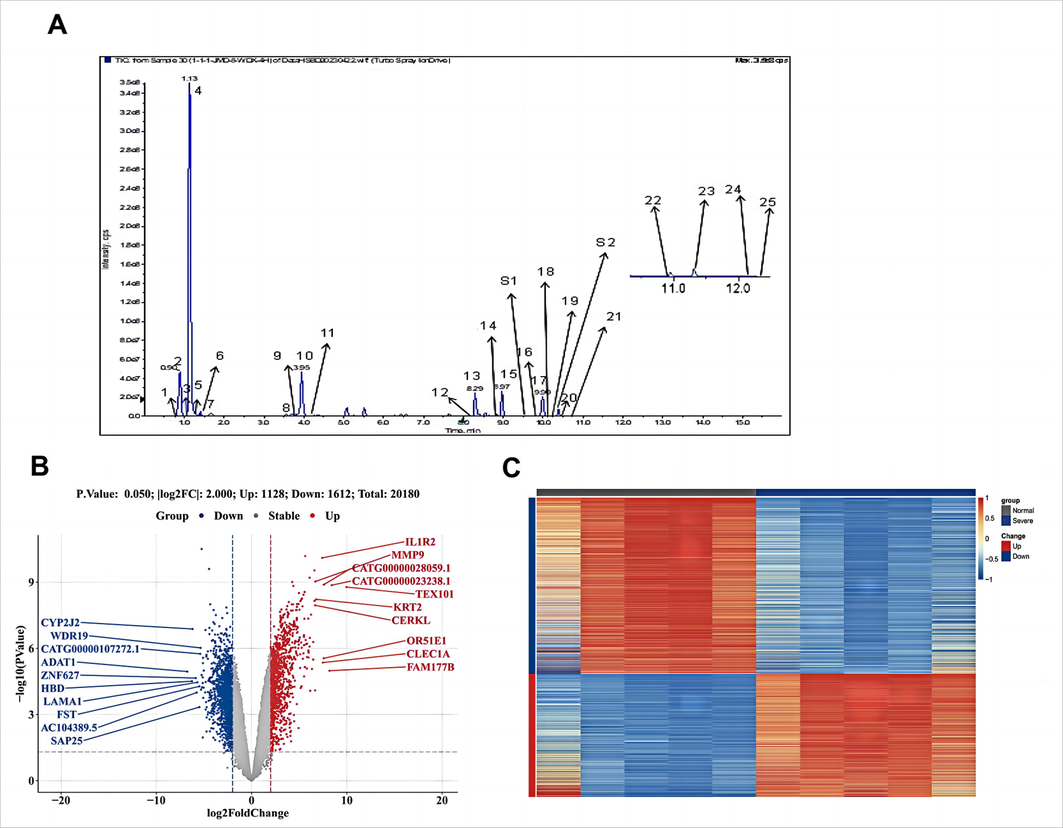
Determination of HSBD sample content components and differential gene acquisition in patients with severe COVID-19. A. MRM profiles of 25 compounds and internal standards in HSBD. 1. Quercetin-3-O-beta-D-glucose-7-O-beta-D-gentiobioside; 2.Isorhamnetin-3-O-beta-D-Glucoside; 3.Pseudoephedrine Hydrochloride; 4.Ephedrine Hydrochloride; 5.Amygdalin; 6. p-Hydroxybenzoic acid; 7.Gentianic acid; 8.Calycosin-7-O-beta-D-glucoside; 9.Hyperoside; 10.Glycyrrhizin; 11.Sinapic acid; 12.Astragaloside; 13.Glycyrrhetinic acid; 14.AloeEmodin; 15.Rhein; 16.Paeoniflorin; 17.Magnolol; 18.Emodin; 19.Honokiol; 20.Pogostone; 21.Hederagenin; 22.Chrysophanic acid; 23.Physcion; 24.Pachymic acid; 25.Atractylon; S1. Cinobufagin; S2. Glibenclamide. B. Differential gene volcano map for COVID-19 severe patients. In this plot, genes are denoted as points, with red indicating up-regulation, blue indicating down-regulation, and gray highlighting no significant difference in expression between severe and normal samples.C. Differential Gene Heat Map for COVID-19 severe patients. Each small square represents each gene, and its color indicates the expression level of the gene, the higher the expression level, the darker the color (red is high expression, blue is low expression). The first row indicates the sample grouping, gray indicates Normal samples and dark blue indicates Severe samples. Each row indicates the expression of each gene in different samples, and each column indicates the expression of all differential genes in each sample.
3.2 Differential gene acquisition in patients with severe COVID-19
The dataset GSE164805 (the transcriptional profiles of severe COVID-19) was retrieved from the Gene Expression Omnibus (GEO) database. The original expression profile dataset contained multiple duplicate symbols. To address this, the median expression for each gene was calculated, and the symbol with the higher median expression was retained (see Appendix: GSE_Matrix.txt). The R package 'limma' was utilized to conduct a differential expression analysis on the refined dataset, comparing severe to normal conditions (see Appendix: GSE_DEGlist.txt). The criteria for differential gene identification were set at an absolute log fold change (|logFC|) greater than 2 and a p-value less than 0.05. This analysis identified 2740 differentially expressed genes, with 1128 being up-regulated and 1612 down-regulated.
To provide a comprehensive overview of the distribution of these differential genes, a volcano plot was generated, as shown in Fig. 1B. Additionally, the expression patterns of these genes are visualized in a heatmap presented in Fig. 1C.
3.3 PPI network of targets of action of HSBD plasma-absorbed components and severe COVID-19 intersecting targets
From the TCMSP database, we identified 429 targets associated with the seven components absorbed into the plasma from HSBD. These targets were cross-referenced with genes related to severe COVID-19, totaling 2,740, resulting in 36 common targets between severe COVID-19 and HSBD plasma-absorbed components(Bardou et al., 2014) (Fig. 2A).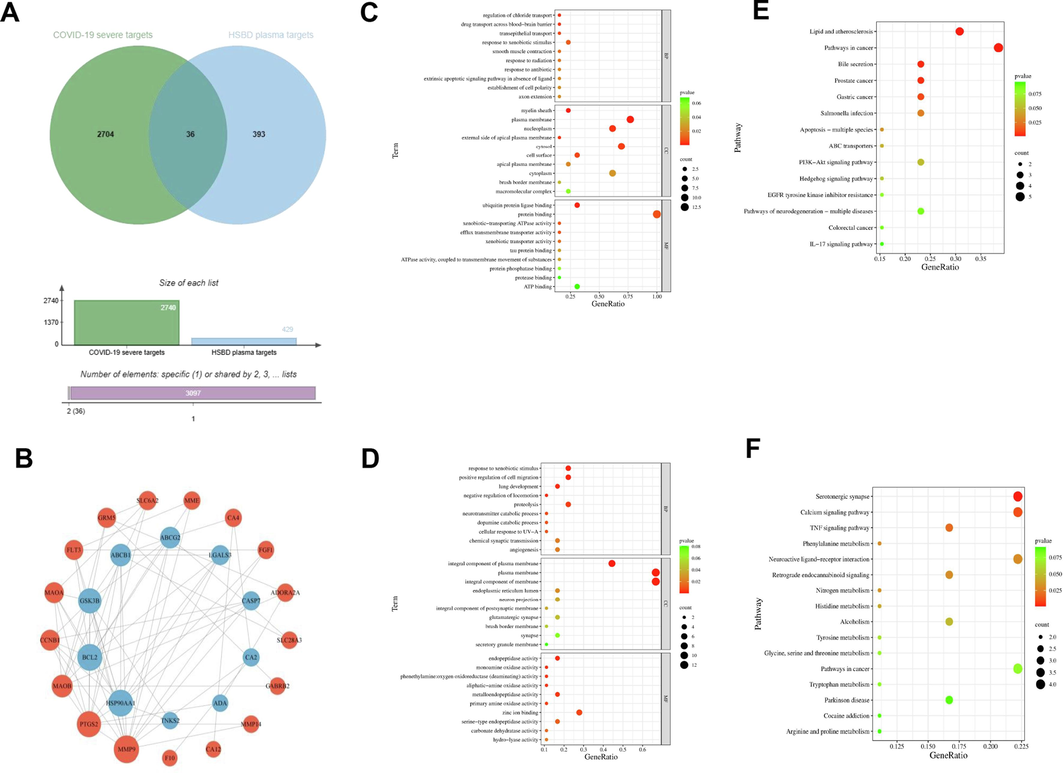
Network pharmacological analysis was conducted to investigate the plasma-absorbed components of HSBD for treating severe COVID-19. A. Identification of shared targets between severe COVID-19 and HSBD plasma-absorbed components. B. PPI network analysis of the action targets for HSBD plasma-absorbed components and their intersection with severe COVID-19 targets. C. GO annotation for down-regulated DEGs. D. GO annotation specifically for up-regulated DEGs. E. KEGG pathway enrichment analysis for own-regulated DEGs. F. KEGG pathway enrichment analysis for up-regulated DEGs.
Utilizing the STRING platform, we constructed a PPI network for the intersecting targets between severe COVID-19 and the HSBD-absorbed components. This network distinguished up-regulated major targets (outer circle) from down-regulated core targets (inner circle). By leveraging data from up-regulated differentially expressed genes (DEGs), we mapped out signaling networks connecting these genes with their corresponding transcription factors. This analysis highlighted key genes and underscored the potential therapeutic implications of active components of HSBD in treating COVID-19. Within this network, the top five down-regulated genes in terms of network connectivity were MMP9, PTGS2, MAOB, CCNB1, and MAOA, whereas the top five up-regulated genes were MAOA, BCL2, GSK3B, ABCB1, and ABCG2 (Fig. 2B). Notably, Matrix Metalloprotein 9(MMP9) emerged as a significant gene, as indicated by its highest connectivity degree, suggesting its crucial role in the anti-COVID-19 response according to the network analysis and corroborative literature.
3.4 GO analysis of core targets
The analysis of down-regulated genes revealed involvement in various biological processes (BPs) such as regulation of chloride transport, drug transport across the blood–brain barrier, and transepithelial transport. Molecular functions (MFs) were associated with structures like myelin sheath and plasma membrane, and cellular components (CCs) included ubiquitin protein ligase binding and xenobiotic-transporting ATPase activity (Fig. 2C).
For up-regulated genes, identified BPs included response to xenobiotic stimulus, positive regulation of cell migration, and lung development. MFs predominantly involved the integral component of the plasma membrane, while CCs feature activities such as endopeptidase and monoamine oxidase activity, highlighted in Fig. 2D. The visualization employed a color and size scheme where redder bubble colors indicated smaller P-values and hence greater significance, while larger bubble sizes denoted a higher number of genes enriched in the corresponding GO entries.
3.5 KEGG pathway enrichment analysis
The KEGG pathway enrichment analysis of upregulated and downregulated differentially expressed genes (DEGs) revealed distinct pathways associated with each gene set. Specifically, the downregulated DEGs were primarily involved in the PI3K-AKT or IL-17 signaling pathways (Fig. 2E), while the upregulated DEGs predominantly influenced the TNF signaling pathway (Fig. 2F).
4 Mechanism of action of the active components of HSBD
Due to market unavailability, Ephedrine hydrochloride and Pseudoephedrine hydrochloride were excluded from the validation process. P-hydroxybenzoic acid, commonly used as an antiseptic, and Pogostone, known for its antimicrobial, anti-inflammatory, antioxidant, and insecticidal properties, were also not included in subsequent validation experiments. Based on the literature and our related studies, Paeoniflorin, Rhein, and Emodin, compounds known to enter the bloodstream, were identified as potentially key active ingredients in HSBD for impacting COVID-19.
In addition to MMP9, combined with reported literature, we selected five additional hot targets targeting COVID-19 to conduct mechanism studies on the active compounds of HSBD. SARS-CoV-2 entry into host cells is characterized by viral spike protein interaction with cellular angiotensin-converting enzyme 2 (ACE2) and human transmembrane protease serine 2 (TMPRSS2) (Berretta et al., 2020). The interaction between a segment of receptor binding domain (RBD) from S protein of the virus and hACE2 is essential for cellular entry of the virus(Nayak, 2021). The viral 3-chymotrypsin-like cysteine protease (3CLpro) and SARS-CoV-2 Papain-Like Protease (PLpro) enzyme controls coronavirus replication and is essential for its life cycle(Nayak, 2021, Amin et al., 2021). MMP9 may be considered as a target for treat severe COVID-19 patients(Gelzo et al., 2022). Therefore, we focused on these six antiviral targets to elucidate the action mechanisms of HSBD active components against COVID-19.
We initially investigated the binding affinities of Paeoniflorin, Rhein, and Emodin with hACE2, RBD, and MMP9 using Surface Plasmon Resonance (SPR) technology. The results, shown in Fig. 3A-3D, and S2, indicated that Paeoniflorin, Rhein, and Emodin all bind to hACE2 and RBD with the dissociation constant (KD) values for hACE2 being 1.9, 8.3, and 31.9 µM and for RBD being 3.4, 13, and 29.1 µM, respectively. Subsequent competition assays were performed to further characterize the binding activity, particularly for Paeoniflorin and Rhein. Notably, at concentrations of 10 and 100 µM, Rhein significantly inhibited the binding to both hACE2 and RBD, while Paeoniflorin exhibited a comparatively weaker inhibition (Fig. 3E and S3). This reduced inhibitory capacity of Paeoniflorin could be attributed to its weaker binding affinity to RBD. Furthermore, SPR assays demonstrated that Paeoniflorin, Rhein, and Emodin could bind to MMP9 with KD values of 21.7, 12.7, and 9.3 µM, respectively (Fig. 3F and S4). However, enzyme activity assays revealed that at 50 µM, none of these three compounds significantly inhibited the activities of 3CLPro, PLPro, and TMPRSS2. These findings suggest that the antiviral effects of Paeoniflorin and Rhein are primarily mediated through their interactions with RBD and ACE2, whereas Emodin's antiviral mechanism is mainly associated with its binding to MMP9.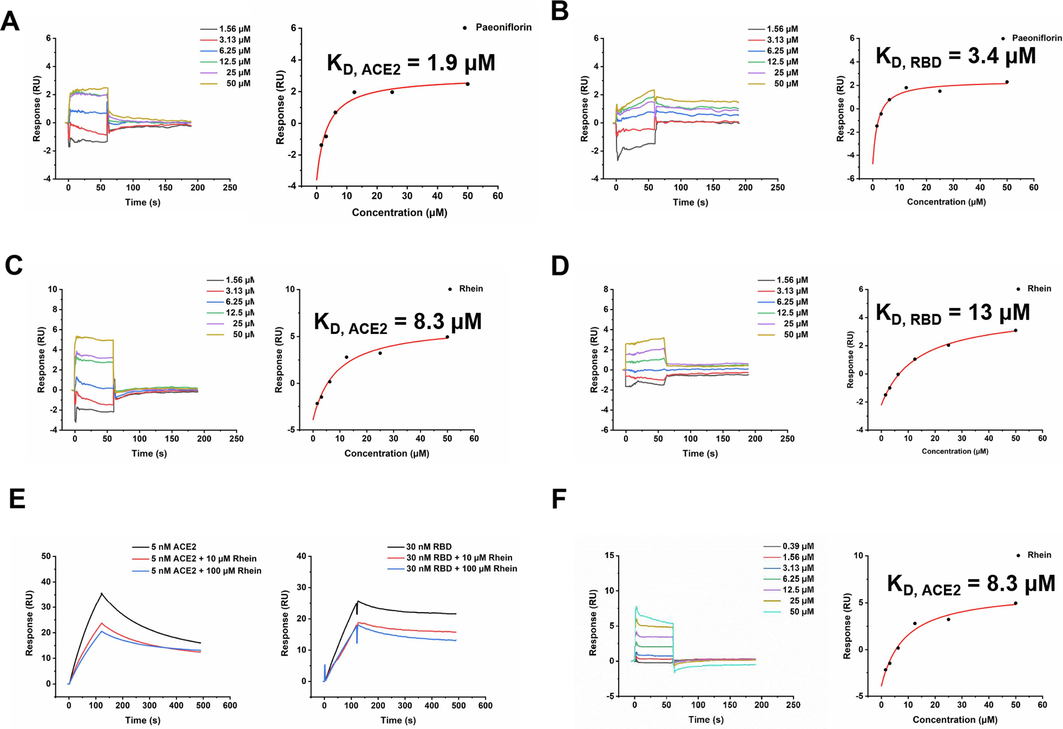
Affinity activity of Paeoniflorin, Rhein, Emodin with hACE2, RBD and MMP9 by SPR technique. Both Paeoniflorin and Rhein bound to hACE2 and RBD, with KD values of 1.9 (A) and 8.3 (C) µM for hACE2 and 3.4 (B) and 13 (D) µM for RBD. A. Competition experiments characterize the binding epitopes of Rhein. It can be seen that Rhein could significantly inhibit the binding of hACE2 and RBD. B. Emodin bound to MMP9 with KD value of 9.3 µM.
5 Molecular docking and dynamics simulation
To elucidate the binding sites of Paeoniflorin, Rhein, and Emodin, molecular docking and molecular dynamics simulations were employed to assess their interactions with RBD, hACE2, and MMP9. Molecular docking results revealed that Rhein and Paeoniflorin have binding energies with hACE2 at −8.673 kcal/mol and −7.891 kcal/mol respectively, and with RBD at −7.079 kcal/mol and −6.727 kcal/mol respectively (Fig. 4A-4B, S5). Specifically, Rhein formed hydrogen bonds with hACE2 at residues GLN98, ASP206, and LYS562. Additionally, its anthraquinone moiety engaged in conjugated interactions with hACE2 at residues LEU95, GLU208, and VAL209. Regarding its interaction with RBD, Rhein established hydrogen bonds with TYR365, LEU390, and CYS391, and exhibited hydrophobic interactions with ALA363 and CYS391. Emodin presented a binding energy of −8.702 kcal/mol with MMP9, while Rhein and Paeoniflorin showed binding energies of −7.869 kcal/mol and −7.542 kcal/mol, respectively (Fig. 4C and S6). Emodin established hydrogen bond interactions with MMP9 at residues TYR48, PRO97, CYS99, and GLU402. Additionally, it demonstrated hydrophobic interactions with residues CYS99, VAL398, PRO421, and TYR423. Concurrently, Emodin engaged in a Pi-lone pair interaction with CYS99, and a Pi-sulfur interaction is observed at the same residue. These findings elucidate the multifaceted interaction dynamics of Rhein with hACE2 and RBD, and Emodin with MMP9, providing valuable insights into its potential inhibitory mechanisms at a molecular level.The binding affinity of Rhein and Paeoniflorin to hACE2, as well as their strong interactions with MMP9 alongside Emodin, were further validated through root mean square deviation (RMSD) and radius of gyration (Rg) analyses. These analyses indicated that the binding of Paeoniflorin to hACE2 stabilized after 30 ns of simulation time, and the binding of Rhein stabilized after 55 ns, suggesting stable and reliable interactions (Fig. 5A and 5B). The radius of gyration results suggested that the binding of these molecules to hACE2 did not significantly alter the overall structural compactness of the protein (Fig. 5C).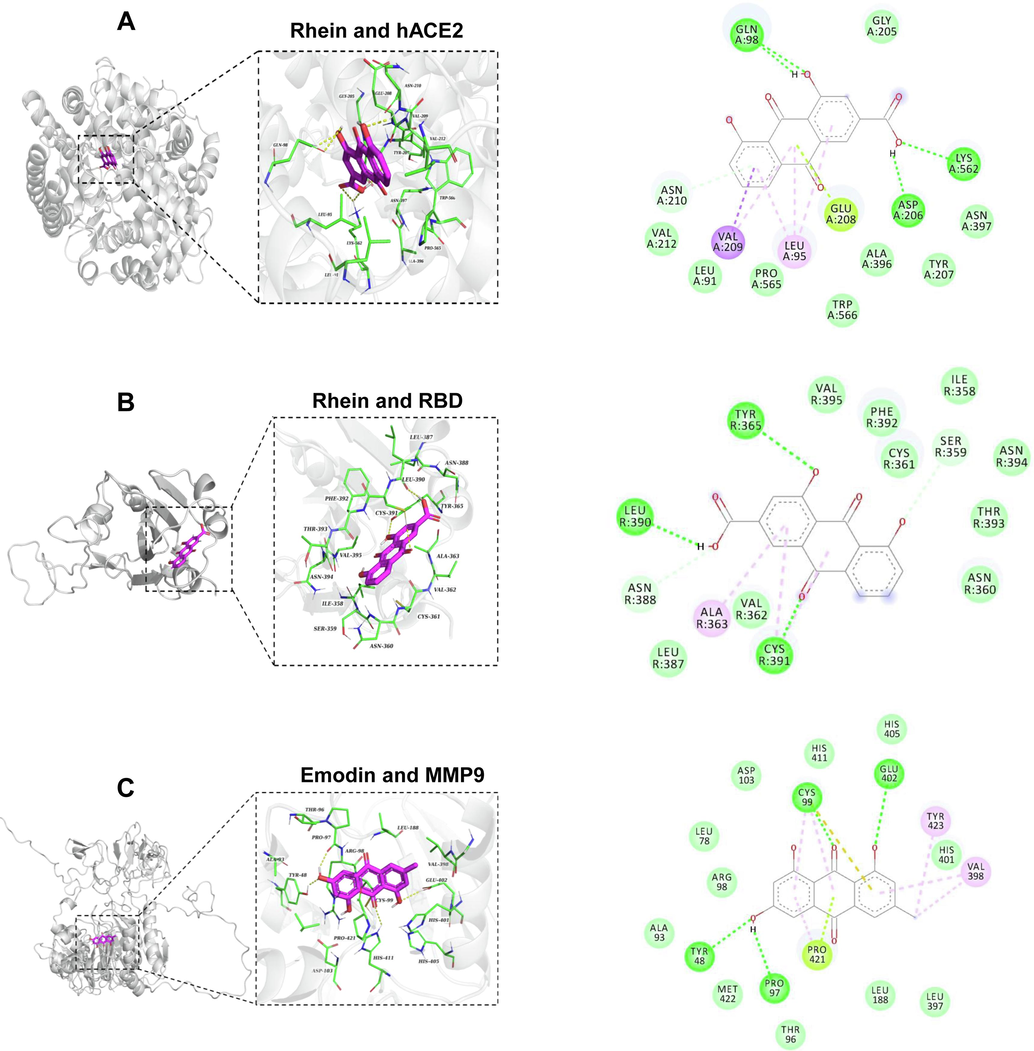
The binding interactions of Rehin or Emodin to ACE2, RBD and MMP9. A. Molecular docking studies revealed the interaction modes of Rhein with hACE2. B. The docking results of Rhein to RBD. C. Molecular docking provided insights into the binding mechanism of Emodin with MMP9 proteins. The dark green dashed lines denote hydrogen bond interactions, whereas the light green, pink, and purple dashed lines depict interactions related to Pi bonds, encompassing both polar and nonpolar interactions.
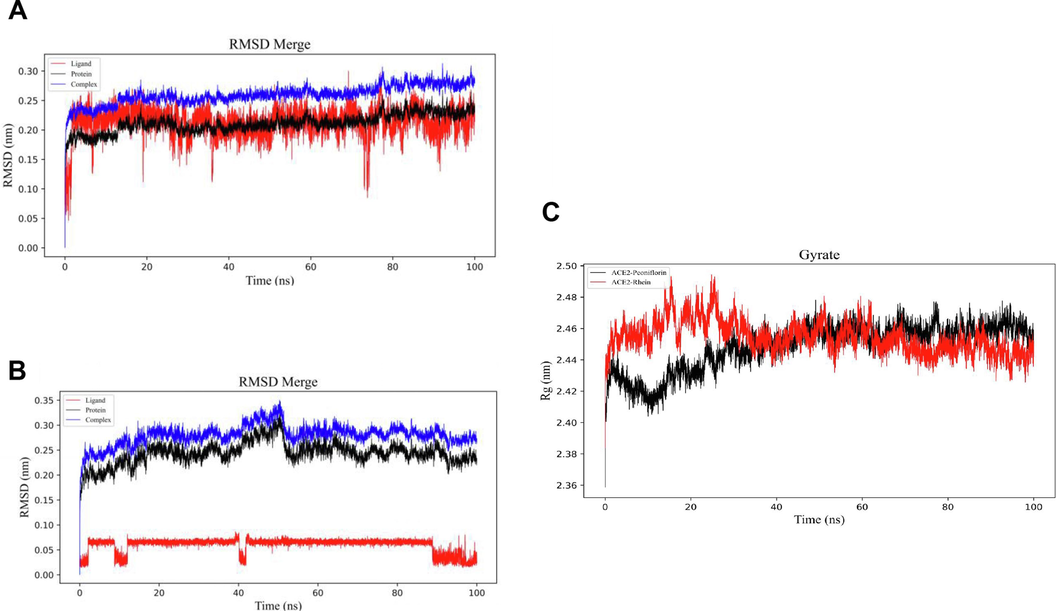
Molecular dynamics (MD) analysis was performed to assess the interaction between Rhein, Paeoniflorin and hACE2, focusing on root mean square deviation (RMSD) and gyration. A. Molecular dynamics simulations of Rhein and hACE2 (RMSD Merge). B. Molecular dynamics simulations of Paeoniflorin and hACE2 (RMSD Merge). C. Molecular dynamics simulations of Rhein, Peoniflorin and hACE2 (Gyrate). Gyrate is an indicator of the overall structural compactness of the protein molecule.
Similarly, the RMSD values for the interactions of Paeoniflorin, Rhein, and Emodin with MMP9 were analyzed, showing stabilization times of 35 ns, 20 ns, and 52 ns, respectively (Figures S6A-S7C). The decreasing trend in the radius of gyration for all three compounds upon binding to MMP9 suggests a more compact overall protein folding (Figures S6D-S6F). These findings provide valuable insights for further research on protein-small molecule interactions and the development of therapeutic agents.
6 Discussion
We employed liquid chromatography-mass spectrometry to validate our methodology and quantify the active ingredients in HSBD, confirming the stability, reliability, and reproducibility of the methods. Among the 25 active constituents of HSBD, seven, including Pogostone, p-Hydroxybenzoic acid, Paeoniflorin, Rhein, Emodin, Ephedrine Hydrochloride, and Pseudoephedrine Hydrochloride, were identified as being absorbed into the bloodstream.
Traditional Chinese Medicine (TCM) syndromes and herbal treatments play a pivotal role in the foundation and evolution of network pharmacology(Wang et al., 2021a). While previous studies on HSBD formula in network pharmacology have focused on the collective targets of the 14 herbs within HSBD and gene acquisition for COVID-19 from GeneCards(Zhu et al., 2021, Zhang et al., 2023), our study introduces a novel approach. We identified the components of HSBD absorbed into the bloodstream using UHPLC-QqQ-MS/MS, enabling quantitative analysis of both major and minor active components in a short time. Additionally, we utilized differentially expressed genes from severe COVID-19 patients, compared to normal subjects, derived from the GEO database, as disease targets.
The dataset GSE164805, related to Chinese patient data, demonstrated HSBD granules' effectiveness in treating COVID-19. Our analysis becomes more meaningful by intersecting the seven identified blood-absorbed HSBD components with severe COVID-19 disease targets for PPI network analysis. This intersection suggests that HSBD could modulate the severity of the disease by downregulating genes like MMP9, PTGS2, MAOB, CCNB1, MAOA, and upregulating genes such as MAOA, BCL2, GSK3B, ABCB1, and ABCG2.
We conducted an in-depth analysis to uncover the potential mechanisms by which the active components of HSBD affect COVID-19, utilizing GO enrichment and KEGG signaling pathway analyses. Our findings indicate that the impact of HSBD on COVID-19 primarily engages the TNF, PI3K-AKT, and IL-17 signaling pathways, all of which are pivotal in the pathogenesis of the disease. TNF, an inflammatory cytokine produced by macrophages/monocytes, plays a critical role in mediating various intracellular signaling processes, including the expression of growth factors, cytokines, and transcription factors. The receptors for TNF are instrumental in modulating the immune response, particularly affecting T-cell activation, proliferation, and survival(Ward-Kavanagh et al., 2016). Furthermore, TNF has been linked to the induction of cytokine storms, a phenomenon closely associated with the severity of COVID-19 due to the resulting immunopathological damage(Ramasamy and Subbian, 2021). Specifically, the upregulation of IL-17A, and potentially IL-17F, within these cytokine storms, contributes significantly to the immunopathology of both COVID-19 and acute respiratory distress syndrome(Shibabaw, 2020). The PI3K-Akt pathway, another crucial cellular signaling route, when activated, can mitigate oxidative harm and inflammatory responses in pulmonary tissue by inhibiting apoptosis, autophagy, or oxidative stress(Matthay and Zimmerman, 2005). Thus, the bioactive compounds in HSBD might potentiate TNF signaling, fostering appropriate immune reactions by upregulating the TNF pathway. This upregulation could enhance TNF expression or its receptor binding, deactivate negative regulators, or activate positive ones, thereby offering therapeutic benefits. Concurrently, HSBD may curb excessive inflammatory responses and prevent tissue damage by downregulating the IL-17 and PI3K-AKT pathways, ensuring a balance between the immune and inflammatory systems. These findings need to be verified through cell and animal experiments in the future.
This study focuses on the research of the components and mechanisms of action of HSBD. To elucidate the action mechanisms of the active constituents in HSBD against COVID-19, in addition to MMP9, we also focused on several viral and human targets: hACE2, RBD, 3CLpro, PLpro, TMPRSS2, employing SPR for our investigations. Our findings revealed that Paeoniflorin and Rhein primarily exhibited antiviral properties through their binding to RBD and ACE2. Conversely, the antiviral mechanism of Emodin predominantly involved interaction with MMP9. Literature supports that MMP9, notably elevated in the lung tissues of COVID-19 patients and integral to cytokine recruitment, plays a crucial role in combating the virus(Hazra et al., 2020). Its upregulation is associated with disease severity, positioning MMP9 as a potential target for assessing COVID-19 severity. Emodin, a natural anthraquinone compound widely found in various plants, possesses anti-inflammatory properties. It can mitigate the excessive inflammatory response caused by COVID-19 infection and reduce the occurrence of cytokine storms by inhibiting the activity of nuclear factor-κB (NF-κB) and other inflammatory mediators(Giovinazzo et al., 2020). Our studies, employing SPR, molecular docking, and molecular dynamic simulation, demonstrated that Emodin effectively targeted MMP9. Therefore, we hypothesized that Emodin may reduce the production of inflammatory factors by inhibiting MMP9 enzyme activity, thereby exerting its antiviral effects, the precise anti-COVID-19 mechanism of Emodin requires further investigation.
However, our study has some limitations. The compounds we identified, including those absorbed into the blood, were based on the active ingredients of traditional Chinese medicines specified in the Pharmacopoeia of the People's Republic of China. Due to these limitations, some potentially effective compounds of HSBD may have been missed. Furthermore, the mechanisms of HSBD and its effective compounds in the treatment of COVID-19 should be further clarified through in vitro and in vivo experiments in the future.
7 Conclusion
In this study, we investigated the methodology and identified the active components of HSBD, focusing on those absorbed into the plasma, to elucidate the effective material basis of HSBD in treating COVID-19. This research shed light on the therapeutic constituents of HSBD and offer insightful exploration into its mechanisms of action against COVID-19.
Authors’ contributions
Chuanxi Tian: Methodology, writing-original draft. Jinyue Zhao: Formal analysis, data Curation. Qian Wang: Methodology, investigation, Data Curation, Writing-Review & Editing, funding acquisition. Keke Luo: Investigation, resources. Shuang Zhao: Writing − Review & Editing. Jiarui Li: Writing − Review & Editing. Li Wan: Writing − Review & Editing. Kaile Ma: Writing −Review & Editing. Yanyan Zhou: Investigation, resources. Min Li: Supervision, project administration, funding acquisition.
CRediT authorship contribution statement
Chuanxi Tian: Writing – original draft. Jinyue Zhao: Investigation, Data curation. Qian Wang: Writing – original draft, Investigation, Data curation. Keke Luo: Investigation. Shuang Zhao: Writing – review & editing. Li Wan: Writing – review & editing. Jiarui Li: Writing – review & editing. Kaile Ma: Writing – review & editing. Yanyan Zhou: Supervision, Investigation. Min Li: Supervision, Project administration, Funding acquisition.
Acknowledgements
This work was supported by National Key Research and Development Program of China (2020YFE0205100) and the National Natural Science Foundation of China (Grants 22107004).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg. Med. Chem.. 2021;29:115860
- [CrossRef] [Google Scholar]
- Propolis and its potential against SARS-CoV-2 infection mechanisms and COVID-19 disease: Running title: Propolis against SARS-CoV-2 infection and COVID-19. Biomed. Pharmacother.. 2020;131:110622
- [CrossRef] [Google Scholar]
- Effectiveness of Chinese medicine formula Huashibaidu granule on mild COVID-19 patients: A prospective, non-randomized, controlled trial. Integr Med Res.. 2023;12:100950
- [CrossRef] [Google Scholar]
- Matrix metalloproteinases (MMP) 3 and 9 as biomarkers of severity in COVID-19 patients. Sci. Rep.. 2022;12:1212.
- [CrossRef] [Google Scholar]
- Can Natural Polyphenols Help in Reducing Cytokine Storm in COVID-19 Patients? Molecules. 2020;25:5888.
- [CrossRef] [Google Scholar]
- Matrix metallopeptidase 9 as a host protein target of chloroquine and melatonin for immunoregulation in COVID-19: A network-based meta-analysis. Life Sci.. 2020;257:118096
- [CrossRef] [Google Scholar]
- Acute lung injury and the acute respiratory distress syndrome: four decades of inquiry into pathogenesis and rational management. Am. J. Respir. Cell Mol. Biol.. 2005;33:319-327.
- [CrossRef] [Google Scholar]
- Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19) Mini Rev. Med. Chem.. 2021;21:689-703.
- [CrossRef] [Google Scholar]
- Critical determinants of cytokine storm and Type I interferon response in COVID-19 pathogenesis. Clin. Microbiol. Rev.. 2021;34:e00299-e00320.
- [CrossRef] [Google Scholar]
- Inflammatory Cytokine: IL-17A Signaling Pathway in Patients Present with COVID-19 and Current Treatment Strategy. J. Inflamm. Res.. 2020;13:673-680.
- [CrossRef] [Google Scholar]
- Efficacy and safety assessment of severe COVID-19 patients with Chinese medicine: a retrospective case series study at early stage of the COVID-19 epidemic in Wuhan, China. J Ethnopharmacol.. 2021;277:113888
- [CrossRef] [Google Scholar]
- TCM network pharmacology: A new trend towards combining computational, experimental and clinical approaches. Chin. J. Nat. Med.. 2021;19:1-11.
- [CrossRef] [Google Scholar]
- The TNF receptor superfamily in co-stimulating and Co-inhibitory responses. Immunity. 2016;44:1005-1019.
- [CrossRef] [Google Scholar]
- Xiong, Y., Tian, Y., Ma, Y., Liu, B., Ruan, L., Lu, C., Huang, L., National Traditional Chinese Medicine Medical Team, 2022. The effect of Huashibaidu formula on the blood oxygen saturation status of severe COVID-19: A retrospective cohort study. Phytomedicine. 95, 153868. 10.1016/j.phymed.2021.153868.
- Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19. PNAS. 2023;120 e2301775120
- [CrossRef] [Google Scholar]
- Efficacy and safety of Huashi Baidu granule plus Nirmatrelvir-Ritonavir combination therapy in patients with high-risk factors infected with Omicron (B.1.1.529): A multi-arm single-center, open-label, randomized controlled trial. Phytomedicine. 2023;120:155025
- [CrossRef] [Google Scholar]
- Huashibaidu formula attenuates sepsis-induced acute lung injury via suppressing cytokine storm: Implications for treatment of COVID-19. Phytomedicine. 2023;109:154549
- [CrossRef] [Google Scholar]
- Coronavirus disease 2019 (COVID-19): a clinical update. Front. Med.. 2020;14:126-135.
- [CrossRef] [Google Scholar]
- Analyzing the potential therapeutic mechanism of Huashi Baidu Decoction on severe COVID-19 through integrating network pharmacological methods. J. Tradit. Complement. Med.. 2021;11:180-187.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2024.105910.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary Data 1
Supplementary Data 1
Supplementary Data 2
Supplementary Data 2







