Translate this page into:
A four-dimensional separation approach by offline 2D-LC/IM-TOF-MS in combination with database-driven computational peak annotation facilitating the in-depth characterization of the multicomponents from Atractylodis Macrocephalae Rhizoma (Atractylodes macrocephala)
⁎Corresponding authors at: State Key Laboratory of Component-based Chinese Medicine, Tianjin University of Traditional Chinese Medicine, 10 Poyanghu Road, Jinghai, Tianjin 301617, China (W. Yang). gaoxiumei@tjutcm.edu.cn (Xiumei Gao), wzyang0504@tjutcm.edu.cn (Wenzhi Yang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Plant metabolites represent complex chemical system, which renders it difficult to clarify the chemical composition by conventional liquid chromatography/mass spectrometry (LC/MS) due to the limited selectivity and peak capacity. The rhizomes of Atractylodes macrocephala have been utilized as a traditional Chinese medicine Atractylodis Macrocephalae Rhizoma (Bai-Zhu), and have been reported containing multiple categories of plant metabolites. Targeting the multicomponents from A. macrocephala, an integral approach by offline two-dimensional liquid chromatography/ion mobility quadrupole time-of-flight mass spectrometry (2D-LC/IM-QTOF-MS) was established and validated. By configuring an XBridge Amide column of Hydrophilic Interaction Chromatography and an Atlantis Premier BEH C18AX column of mixed ion exchange and reversed-phase modes, the established 2D-LC/IM-QTOF-MS system showed high orthogonality up to 0.91. Dimension-enhanced, data-independent high-definition MSE (HDMSE) in the positive ESI mode was conducted on a Vion IM-QTOF mass spectrometer, and its hyphenation to offline 2D-LC could enable the four-dimensional separation (each dimension in 2D-LC, IM, and MS). Particularly, HDMSE facilitated the acquisition of high-definition MS1 and MS2 spectra. In-house library-driven computational peak annotation by the bioinformatics platform UNIFI could efficiently process and annotate the HDMSE data for the structural elucidation. By integrating reference compounds comparison, we could identify or tentatively characterize 251 components from A. macrocephala (including 115 sesquiterpenoids, 90 polyacetylenes, 11 flavonoids, 9 benzoquinones, 12 coumarins, and 14 others), which indicated large improvement in identifying those minor plant components, compared with the conventional LC/MS approach. Conclusively, offline 2D-LC/IM-QTOF-HDMSE in combination with computational data interpretation proves to be powerful facilitating the in-depth multicomponent characterization of herbal medicine.
Keywords
Two-dimensional liquid chromatography
Ion mobility quadrupole time-of-flight mass spectrometry
Multicomponent characterization
In-house database
Atractylodes macrocephala
1 Introduction
As an important resource for the medicine development, traditional Chinese medicine (TCM) has been included in the system of alternative therapy and is attracting the increasing attention from a global scope. However, there are several obstacles hindering the internationalization process of TCM. TCM, especially the herbal medicine, has complex chemical compositions (the co-existing of the primary and secondary metabolites featured by the wide spans of acidity-base property, polarity, molecular mass, and content, etc.), which raises great challenges for performing the quality investigation (Yang et al., 2017). Advances in analytical technologies have largely driven the profiling and characterization of versatile natural compounds from TCM and the compound formulae, of which liquid chromatography/mass spectrometry (LC/MS) currently is the most preferable in the untargeted and targeted qualitative analysis of TCM components, as multiple mechanism of chromatography and personalized MS scan strategies have been developed (Ganzera and Sturm, 2018; Saurina and Sentellas, 2019; Stavrianidi, 2020). On the one hand, the accessibility of reversed-phase chromatography (RPC), hydrophilic interaction chromatography (HILIC), ion exchange chromatography (IEC), size exclusion chromatography (SEC), and even the ongoing developing supercritical fluid chromatography (SFC), almost can cover all the known types of plant metabolites with good resolution (Jin et al., 2016; Paczkowska et al., 2019). On the other hand, diverse scan methods (including the data-dependent acquisition-DDA and data-independent acquisition-DIA) and fragmentation mechanisms (collision-induced dissociation-CID, high-energy collision-induced dissociation-HCD, pulsed-Q dissociation-PQD, and electron transfer dissociation-ETD, etc.) can be alternative for use to profile and characterize the multicomponents from TCM. In spite of these favorable merits, the application of one-dimension separation based LC/MS to deconvolute the chemical complexity of TCM is still encountered with several insufficiencies: i) the selectivity of separation and peak capacity of chromatography are limited, which causes the poor resolution of those easily co-eluting components and limited exposure of the minor ingredients that are easily covered by the rich ones; ii) the low coverage on target components by conventional DDA approaches or the lack of efficient tools to process the DIA data, which always results in unsatisfactory characterization results; iii) short of specific library for the TCM of study; and iv) defective ability to differentiate isomers. These issues can, to some extent, lead to insufficient separation and compromised performance in identifying the multicomponents, which thus necessitates to develop more powerful analytical approaches in support of the comprehensive multicomponent characterization of TCM.
Benefitting from the development of multi-dimensional liquid chromatography (MDLC) and the outstanding resolution, sensitivity, and alternative scan methods of modern MS instruments, the researchers can largely elevate the performance of LC/MS in TCM component characterization, by which hundreds of components can be identified from a single herb sample (Qiao et al., 2016; Qiu et al., 2015; Yao et al., 2018). First, two-dimensional liquid chromatography (2D-LC), in recent five years, have covered more and more applications in the multicomponent characterization of TCM as well as the compound formulae, which is able to separate and identify much more compounds than conventional LC/MS due to the inclusion of twice chromatographic separation, in particular using the orthogonal chromatography (Pirok, Stoll and Schoenmakers, 2019; Zhou et al., 2020). 2D-LC can run in either an offline or an online mode, and each has its merits and demerits. Comparatively, online 2D-LC is highly automatic and easier to standardize (Cao et al., 2019; Zhang et al., 2018), while the offline mode is advantageous for its easy accessibility and high flexibility in integrating different mechanism of chromatography to achieve the maximum orthogonality (Fu et al., 2018; Pan et al., 2018). To date, the scientists are free to perform a systematic chemical analysis experiment by comprehensive 2D-LC or target qualitative/quantitative determination of those interested ingredients by heart-cutting 2D-LC. Second, more potent MS scan strategies are developed to target more components from a given TCM sample, which is impressed by three characteristics: i) the inclusion of a precursor ions list or an exclusion list in DDA (Li et al., 2020a; Pan et al., 2020); ii) the application of novel DIA approaches, such as SWATH (Sequential Windowed Acquisition of All Theoretical Mass Spectra for SCIEX AB QTOF) and in-house establishment of algorithm to efficiently process the obtained DIA data (Xia et al., 2019; Yin et al., 2019); and iii) the development of information-dependent acquisition/enhanced product ions (IDA-EPI) scan methods, such as pMRM-IDA-EPI or MIM-IDA-EPI, etc. (Li et al., 2019; Marek Dziadosz, 2019; Yan et al., 2014). Another important advancement in TCM component analysis in recent years is the introduction and ongoing application of ion mobility mass spectrometry (IM-MS), which provides more possibilities to address the challenging separation encountered in TCM analysis. IM-MS is capable of differentiating the ions on the basis of the charge state, size, and shape, which is orthogonal to the separation by m/z values. Particularly, when coupled with ultra-high-performance liquid chromatography (UHPLC), the use of IM-MS can obtain four-dimensional data of the analytes, including the retention time (tR), drift time, MS1/MS2 information, and the response (Li et al., 2020b; D’Atri et al., 2018). More importantly, IM separation can give the information of collision cross section (CCS), a stable physical parameter among different labs or on different instruments, which can be utilized for creating database beneficial to the metabolites identification and even the isomers differentiation (Tu et al., 2019; Zhou et al., 2017, 2016). By using the VionTM IM-QTOF LC/MS platform, we recently established the IM-enabled high-definition MSE (HDMSE) approach combined with standardized data processing workflows using the bioinformatics software UNIFI (Waters), which has been proven as efficient and powerful in the multicomponent characterization and multi-batch metabolomics analysis for the quality control of TCM (Jia et al., 2019; Koley et al., 2020; Wang et al., 2020; Zhang et al., 2019; Zuo et al., 2019).
The rhizomes of Atractylodes macrocephala have been used as a traditional Chinese medicine Atractylodis Macrocephalae Rhizoma (Bai-Zhu), for the treatment of spleen hypofunction, loss of appetite, abdominal distension, diarrhea, dizziness, and heart palpitation, etc. (Zhu et al., 2018). Phytochemical researches have reported the presence of volatile oils, polysaccharides, lactones, flavonoids, and amino acids, etc., among which atractylodes lactones (e.g. atractylenolides I, II, and III) are a class of sesquiterpenoids exerting the pharmacological properties. Pharmacological studies showed that, A. macrocephala has the functions of anti-tumor, anti-inflammatory, regulating gastrointestinal movement, and regulating immunity (Dong et al., 2008; Kou et al., 2017; Li et al., 2014; Xu et al., 2012). At present, although there have been reports regarding the quality investigations of A. macrocephala (Meng et al., 2019; Shan et al., 2014; Shi et al., 2012; Sun et al., 2017), currently available LC/MS approaches can only characterize tens of compounds from the rhizome, making those minor ones failing to be exposed and identified (Shadab et al., 2020). More powerful analytical technology, by focusing on the enhancement of chromatographic separation and the coverage of MS detection, should be developed.
In this work, we were aimed to develop an offline two-dimensional liquid chromatography/ion mobility quadrupole time-of-flight mass spectrometry (2D-LC/IM-QTOF-MS) approach combined with in-house library-driven automatic peak annotation using UNIFI, for the in-depth profiling and characterization of the multicomponents from A. macrocephala. The data-independent HDMSE in the positive ESI mode was employed for data acquisition. Especially, due to the enabling of four-dimensional separations (2D-LC, IM, and MS), the resolution and characterization of A. macrocephala components could be largely enhanced. To standardize the method development, the approaches used to construct, optimize, and evaluate the performance of the 2D-LC/IM-QTOF-MS approach were offered. The UNIFI workflows by searching an in-house database that could facilitate the automatic annotation of the obtained multiple HDMSE data (corresponding to the fractionated samples of A. macrocephala) were established. Comparison with the reference compounds and fragmentation pathways analysis, together with the retrieval of the self-built library, were utilized to re-analyze and confirm the software-aided characterization results, as a consequence, a total of 251 components were identified or tentatively characterized, which, in contrast to the method reported in literature (251 VS 52) (Sun et al., 2017), indicated large improvement by 4.83 folds in identifying the multicomponents from A. macrocephala.
2 Materials and methods
2.1 Reagents and chemicals
In total, nineteen compounds (Fig. S1 and Table S1), involving five terpenoids, three phenylpropanoids, two volatile oils, one oligosaccharide, three flavonoids, and five others, were purchased from Shanghai Standard Biotech. Co., Ltd. (Shanghai, China) and Chengdu Desite. Co., Ltd. (Chengdu, China), and used as the reference compounds in this work. Acetonitrile, methanol (Fisher, Fair lawn, NJ, USA), formic acid (FA), and (ammonium acetate) (AA; Fisher, Fair lawn, NJ, USA) were LC-MS grade. Ultra-pure water was in-house prepared using a Milli-Q Integral 5 water purification system (Millipore, Bedford. MA, USA). The sample of A. macrocephala analyzed in this work was collected from Pan’an of Zhejiang Province. The voucher specimen of A. macrocephala was deposited in the authors’ lab in Tianjin University of Traditional Chinese Medicine (Tianjin, China).
2.2 Sample preparation
To the sample of A. macrocephalas, 6.0 g of the accurately weighed powder was soaked in 10 mL of 70% aqueous methanol (v/v), vortexed for 2 min, and extracted by ultrasound assistance for 40 min in a water bath at 25 °C. After a 10-min centrifugation at 3,219 g, the lost weight was compensated with 70% methanol. The supernatant was taken and further diluted in a 10-mL volumetric flask to the constant volume. Suffering from another centrifugation process at 11,481 g for 10 min, the supernatant obtained was used as the test solution (with the final concentration at 600 mg/mL).
2.3 Offline 2D-LC/IM-QTOF-MS
Combination of HILIC × RPLC was used to establish an offline 2D-LC system for separating the chemical components contained in A. macrocephalas. In detail, the total extract of A. macrocephalas was initially separated by an XBridge® Amide column (4.6 × 150 mm, 3.5 µm) on an Agilent 1260 HPLC system (Agilent Technologies, Waldbronn, Germany). The column temperature was maintained at 30 °C and the binary mobile phase consisting of 0.1% formic acid (A) and acetonitrile (B) at a flow rate of 1.0 mL/min was used following a gradient elution program: 0–2 min, 98%–90% (B); 2–6 min, 90%–85% (B); 6–8 min, 85%–80% (B); 8–10 min, 80%–75% (B); 10–11 min, 75%–70% (B); 11–12 min, 70%–67% (B); 12–14 min, 67%–65% (B); 14–14.5 min, 65%–50% (B); and 14.5–16 min, 50% (B). The injection volume was 40 µL. The injection was repeated for five times. The PDA detector monitored the signals at 282 nm. Eluate collection was based on a peak-dependent fractionation approach, which finally gave eight fractions (numbered as Fr. 1 to Fr. 8). Each of the dry residues corresponding to eight fractions was dissolved in 200 µL of 70% methanol (v/v) and further centrifuged at 11,481 g for 10 min, the resultant supernatants were the test solutions (with the final concentration at 1.0 mg/mL equal to the raw drug material) for the second-dimensional UHPLC separation in the RP mode. An Atlantis Premier BEH C18AX column (2.1 × 100 mm, 1.7 µm) maintained at 30 °C was configured on an ACQUITY UPLC I-Class/Vion IM-QTOF system (Waters Corporation, Manchester, UK) for the 2D separation. A binary mobile phase, containing 0.1% formic acid (A) and acetonitrile (B), ran consistent with the following gradient program: 0–3 min, 3%–30% (B); 3–6 min, 30%–50% (B); 6–10 min, 50%–60% (B); 10–13 min, 60%–65% (B); 13–16 min, 65%–75% (B); 16–19 min, 75%–85% (B); 19–24 min, 85%–93% (B); 24–24.5 min, 93%–98% (B); and 24.5–26.5 min, 98% (B). The flow rate was kept at 0.3 mL/min, and an injection volume of 10 µL was used.
High-accuracy MS data were acquired on a Waters Vion™ IM-QTOF mass spectrometer (Waters, Manchester, UK) coupled with the UPLC I-Class system via a Zspray™ ESI source. In order to obtain more reliable characterization results, high-definition MSE in positive mode was utilized. The ESI source parameters were set as follows: capillary voltage, 3.0 kV; cone voltage, 60 V; source offset voltage, 80 V; source temperature, 120 °C; desolvation temperature, 500 °C; cone gas flow rate, 50 L/h; and desolvation gas flow rate, 800 L/h. For HDMSE settings, the mass analyzer scanned over a mass range of 50–1400 Da in full scan with a scan time of 0.3 s under the low collision energy of 6 eV. MS2 experiments were performed by collision-induced dissociation (CID) at the ramp collision energy (RCE) of 20–50 eV covering the same scan range as MS1. Data calibration was performed using an external reference (LockSpray™) by the constant infusion of the leucine-enkephalin solution (LE; Sigma-Aldrich; 200 ng/mL) at a flow rate of 10 μL/min. Calibration of CCS was conducted according to the manufacture’s guidelines using a mixture of calibrants (Paglia et al., 2015).
2.4 Orthogonality and peak capacity
Orthogonality of the developed offline 2D-LC/IM-QTOF-MS system was assessed by the asterisk equations proposed by Camenzuli and Schoenmakers (Camenzuli and Schoenmakers, 2014). These equations are provided as Supplementary Materials. Retention time for each of the 54 index components in both two dimensions of separations was firstly transformed into the normalized retention time (tR,norm(i)) following Eq. (1) (tD: dead time; tG: total gradient elution time). The separation space could be described by four crossing lines (Sz-, Sz+, Sz1, Sz2), and the spreading of all 54 index components around them was quantified by Eqs. (2)–(5) (σ stands for the standard deviation). Four Z parameters (Z-, Z+, Z1, Z2) were then generated in accordance with Eqs. (6)–(9), based on which orthogonality (A0) was determined using Eq. (10).
2.5 Establishment of the in-house library of A. macrocephalas
An in-house library of A. macrocephalas was created by giving a comprehensive summary on the compounds ever isolated from this species. The literature from Web of Science (www.webofknowledge.com), CNKI (www.cnki.net), ChemSpider (www.chemspider.com), PubChem (http://pubchem.ncbi.nlm.nih.gov), and Chemicalbook (www.chemicalbook.com), etc., dealing with the phytochemical isolation of A. macrocephalas was retrieved. First, according to the information provided in the literature, the structure of each compound was prepared by the ChemDraw Professional software, which was saved as an .mol file. Second, the structural information of all known compounds was input into the EXCEL file in a prescribed format. The .mol file was named with the trivial name consistent with that in the EXCEL file. Third, the EXCEL file and all the structure files were incorporated into the UNIFI™ software through Import Library Item, which could be applied as a database for the subsequent peak annotation. A table detailing the in-house library of A. macrocephalas has been provided as Table S2.
2.6 Automatic peak annotation of the HDMSE data by UNIFITM for structural elucidation
Structural elucidation of the multicomponents from A. macrocephalas was performed following an efficient computational workflow. The UNIFI software was applied to process and annotate the collected HDMSE data (of eight fractionated samples) by searching the incorporated in-house library of A. macrocephalas. After corrected by reference to the LockMass at m/z 556.2766 (the theoretical m/z value for LE in the ESI+ mode), a list of the characterized components was generated, with the retention times and aligned characteristic fragments. Adduct ions filtering and MS2 data analysis were further utilized to remove the false positive characterization results and confirm the identities.
The uncorrected HDMSE data in Continuum were further processed using UNIFI 1.9.3.0 (Waters). The UNIFI software performed data correction, peak picking, and peak annotation efficiently. Key parameters set in UNIFI were as follows. Find 4D Peaks: High-energy intensity threshold, 500.0 counts; low-energy intensity threshold, 1000.0 counts. Target by mass: Target match tolerance: 10.0 ppm; screen on all isotopes in a candidate, generate predicted fragments from structure, and fragment match tolerance, 10.0 ppm. Adducts: Positive adducts including +H, +Na. Lock Mass: Combine width, 3 scans; mass window, 0.5 m/z; reference mass, 556.2766; reference charge, +1.
3 Results and discussion
3.1 Development of an offline 2D-LC/IM-QTOF-MS system enabling the four-dimensional separation for the multicomponent profiling and characterization of A. macrocephala
Good chromatographic separation is a prerequisite to comprehensively identify the multicomponents in a complicated chemical system like an herbal extract. Conventional LC/MS approaches, due to the utilization of a single mechanism of separation (such as the most preferable RPC) may suffer from severe co-eluting which largely restrains the exposure of those minor ingredients. Herein, we propose to establish a four-dimensional separation approach, by offline 2D-LC/IM-QTOF-MS, to tackle the insufficiency encountered in the comprehensive characterization of TCM components. By this method, 2D-LC can enable orthogonal chromatographic separation with significantly improved peak capacity, and IM-QTOF-MS renders orthogonal IM and MS separations providing the CCS and high-resolution MS1/MS2 information.
The offline 2D-LC system was optimized, in the first step, by screening the stationary phase and optimizing the column temperature and gradient elution program in each dimension of chromatography (Fu et al., 2018; Qiu et al., 2015; Yao et al., 2018). Considering the integration of different separation mechanisms can improve the selectivity and promote the exposure of the minor components from TCM, efforts were made to achieve large orthogonality of two-dimensional separations (Stavrianidi, 2020). Meanwhile, taking into account the high selectivity and the compatibility with the mass spectrometer of the RP mode, RPC was selected as the second-dimensional (2D) chromatography coupled with MS. In this section, eight candidate sub-2 µm particles packed columns featured by different silica gel core, different bonding groups and bonding technologies, including BEH C18, Zorbax Extend C18, Zorbax SB C18, Atlantis Premier BEH C18AX, HSS T3, Zorbax Eclipse Plus C18, Kinetex XB-C18, and Luna Omega Polar C18, were tested using a total extract of A. macrocephala. In general, the resolution of the multicomponents from A. macrocephala by these columns showed significant difference. Comparatively, BEH C18 (the resolvable ions: 10184), Zorbax SB-C18 (10188), Atlantis Premier BEH C18AX (10173), Zorbax Eclipse Plus C18 (10187), and Luna Omega Polar C18 (10184), were able to resolve more peaks than the other three (Fig. 1A), showing better selectivity. By further comparison of these five columns mentioned, we could find that Atlantis Premier BEH C18AX exhibited much better peak symmetry and column efficiency than the others, which was thus considered as the best choice in the 2D chromatography. It is noted that, Atlantis Premier BEH C18AX is a mixed mode column newly released by Waters, which enables the separation of complex ingredients simultaneously by the RP and ion exchange modes. The combined separation modes lead to good resolution of the multicomponents from A. macrocephala. In the next step, by using 19 compounds as the index components, the linearity regression correlation coefficient (R2) was used as a parameter to compare the separation difference between another six candidate columns with Atlantis Premier BEH C18AX that had been selected in the 2D chromatography (Fu et al., 2018). Notably, these candidate columns have different separation mechanisms (RP and HILIC) or are bonded with different functional groups. As exhibited in the right part of Fig. 1A, we could primarily deduce the separation difference with the Atlantis Premier BEH C18AX mixed mode column was consistent with the order: Zorbax HILIC Plus > BEH HILIC > XBridge Amide > COSMOCORE PBr > Exsil Plus 100 C18-PFP > XCharge C18, based on the increasing R2 values. However, the Zorbax HILIC Plus and BEH HILIC columns were considered unsuitable for separating the multicomponents of A. macrocephala because of the poor retention ability. As a result, the XBridge Amide column was finally selected as the first-dimensional (1D) column. The column temperature and gradient elution program for each dimension of chromatography were optimized as well, which resulted in the establishment of the offline 2D-LC system achieving sufficient resolution of the multicomponents from A. macrocephala.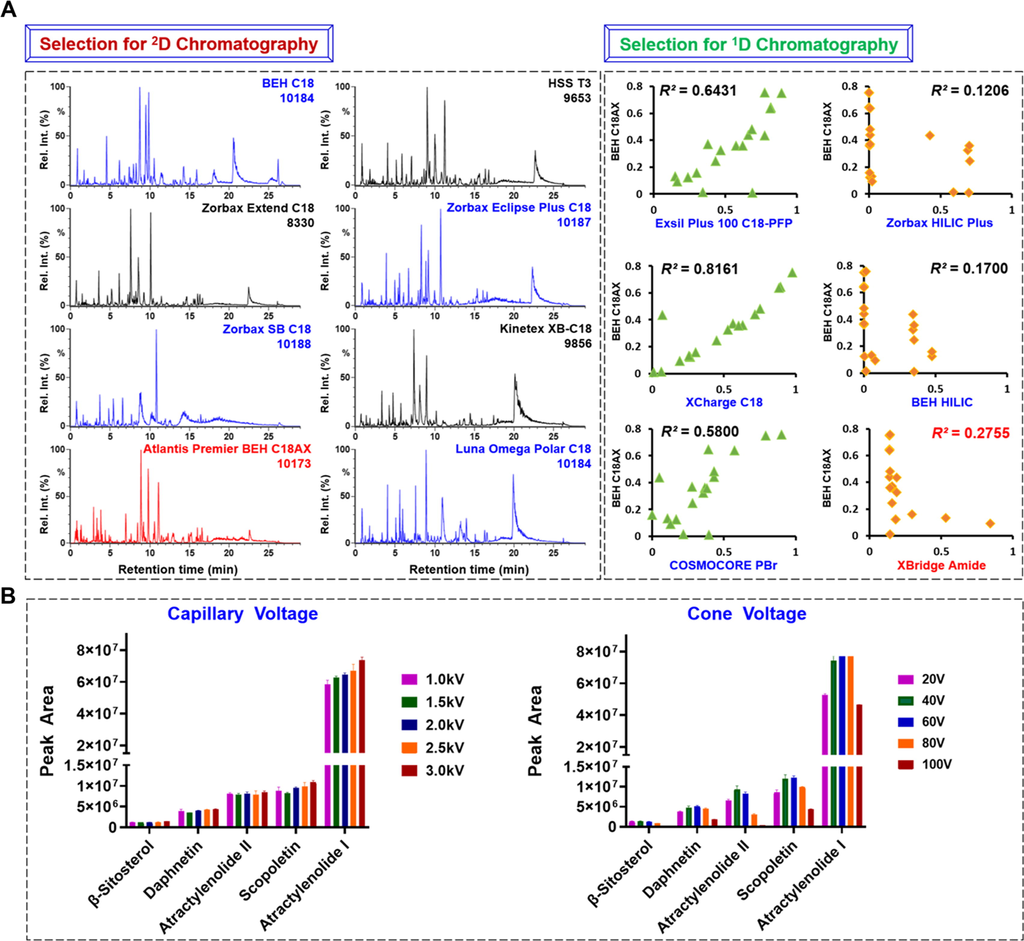
Selection of the stationary phase in establishing an offline 2D-LC system (A) and optimization of two key source parameters (capillary voltage and cone voltage) of the Vion IM-QTOF mass spectrometer (B) dedicated to well separating the multicomponents from Atractylodes macrocephala. Left column of section A presents the base peak intensity (BPI) chromatograms of eight candidate columns in the 2D chromatography together with the number of resolvable peaks; right column is the scattering plots of index components based on the relative retention time (ranging from 0 to 1) determined on six candidate columns in 1D chromatography (x-axis) and the selected Atlantis Premier BEH C18AX column of 2D separation (y-axis). R2 stands for the linearity regression correlation coefficient, and a smaller value can indicate large separation difference between the two columns. The histograms of section B are plotted by the peak areas of the precursor ions for five representative compounds determined at five different levels.
Key parameters of the Vion IM-QTOF instrument operating in the positive ESI mode, involving two key ion source parameters (capillary voltage and cone voltage) and the ramp collision energy (RCE), were further optimized by comparing the variations of peak areas of the precursor ions for five representative components (involving β-sitosterol, daphnetin, atractylenolide II, scopoletin, and atractylenolide I). Capillary voltage and cone voltage can affect the ion response and may induce in-source decay. For capillary voltage varying between 1.0 and 3.0 kV, the peak areas for most of the index components positively correlated with the increasing of capillary voltage, and the setting of 3.0 kV could yield the best ion response and good repeatability through the triplicate operations. Histograms showing the variation tendencies of the ion response for five index components among five different levels of cone voltage (20–100 V) are given in Fig. 1B. A clear “increasing and decreasing” trend was observed, and the best ion response was obtained at cone voltage of 40 V or 60 V. As the highest precursor ion peak areas for more components could be obtained at cone voltage 60 V, in this work, we set the cone voltage in the positive mode at 60 V. In addition, the quality of the MS2 spectra and the number of characterized components largely depend on the collision energy (Saurina and Sentellas, 2019). The Vion IM-QTOF mass spectrometer enables RCE, which can benefit the obtaining of more balanced MS2 spectra (Jia et al., 2019; Zhang et al., 2019; Zuo et al., 2019) than the single collision energy. We set four groups of RCEs, including 10–40 eV, 20–50 eV, 30–60 eV, and 40–70 eV with an increase of 30 eV in each collision ramp, to compare the CID-MS2 data of atractylenolide I, atractylenolide II, atractylon, biepiasterolide, and 8-β-ethoxyasterolid, as the representative compounds (Fig. S2). We could discern a remarkable tendency that increasing of RCE enabled more fragmentation of the precursors yielding more diversified product ions with medium mass, and then more low-mass product ions with higher intensity were obtained when RCE further ascended. To simultaneously retain the precursor ions and obtain rich fragments, we considered the choice of RCE 20–50 eV in this study.
3.2 Orthogonality, peak capacity and method validation for the established offline 2D-LC/IM-QTOF-MS system
Peak capacity and orthogonality are the core indicators for evaluating the separation performance of a 2D-LC system (Pirok, Stoll and Schoenmakers, 2019). In this work, orthogonality evaluation was based on a series of asterisk equations established by Camenzuli and Schoenmakers by using 54 index components (Camenzuli and Schoenmakers, 2014). Distribution degree of these compounds in a two-dimensional plane could be depicted by four lines (Fig. S3). The results showed the selected compounds were relatively evenly distributed around these four lines, with Z-, Z+, Z1, and Z2 calculated at 0.99, 0.93, 0.92, and 0.98, respectively, based on which orthogonality (A0) of the established 2D-LC system was 0.91. In addition, the average peak widths at the baseline in 1D (HILIC) and 2D (mixed mode of IEC and RPC) chromatography were 0.30 min and 0.19 min, respectively, and accordingly, we could deduce the peak capacity of 66 for 1nc and 157 for 2nc. Thereby, the theoretical peak capacity of this 2D-LC system (n2D) was 10362. The high orthogonality between the selected two dimensions of chromatography and largely improved peak capacity can testify the potency of the elaborated 2D-LC system, and thus lay foundation for the separation and identification of more components from A. macrocephala, in particular those minor ones.
To render a powerful analytical approach potentially with wide application, method validation was performed in terms of the intra-day/inter-day precision, stability, repeatability, and limit of detection (defined for an analyte at the lowest concentration the fragments generated can be used for structural characterization), considering its qualitative analysis property (Fu et al., 2018; Yang et al., 2016). Precision was evaluated by six consecutive injections on the first day and another three injections separately on the second and third days. The intra-day and inter-day precision varied 0.98–3.22% and 1.91–5.03% for the 1D separation, and 1.01–5.00% and 3.82–13.03% for the 2D separation, respectively. Repeatability was assessed by analyzing six reduplicative samples of the sub-sample Fr. 1 (the first fractionation sample after the 1D HILIC separation), and repeatability was less than 15% among six samples (Table S3–S5). LOD for six reference compounds (atractylenolide I, atractylenolide II, luteolin, scopoletin, β-sitosterol, apigenin) varied between 0.33 and 0.44 ng. The obtained results could indicate the sensitive and stabilized performance of the configured 2D-LC system suitable for the qualitative analysis of the multicomponents from A. macrocephala.
3.3 Comparison between HDMSE and MSE
The use of HDMSE on the Vion IM-QTOF mass spectrometer offers high-definition full-scan spectrum. We compared the performance between the IM-enabled HDMSE and IM-disabled MSE in characterizing the multicomponents from the extract of A. macrocephala. Consequently, the differentiations in the data obtained by these two methods can be reflected in two aspects. First, the ion response for the components of A. macrocephala was significantly different, and the absolute abundance recorded by MSE was much higher than that determined by HDMSE (Fig. S4). For instance, the peak area of atractylenolide II determined by MSE was 7.3 folds of that obtained by HDMSE. We are aware that the additional IM separation can separate the ions with the same m/z but different charge state or shape, which may result in the reduction in ion response recorded by HDMSE. Second, because of the enhanced separation ability facilitated by HDMSE, the composition of ion species in full-scan MS1 spectra became simpler (dubbed high-definition spectra), in contrast to MSE. As shown in Fig. 2B, three components (tR at 6.96, 10.95, and 12.99 min) got better resolved by HDMSE showing much less interference. HDMSE enabled the additional IM separation providing one new dimension of drift time, compared with MSE. Accordingly, the CID-MS2 spectra of single compounds obtained by HDMSE (especially for atractylenolide II) seemed cleaner, making the data interpretation work easier to implement (Fig. S5). Because of these merits, we selected HDMSE to acquire the fragmentation information of the components from A. macrocephala.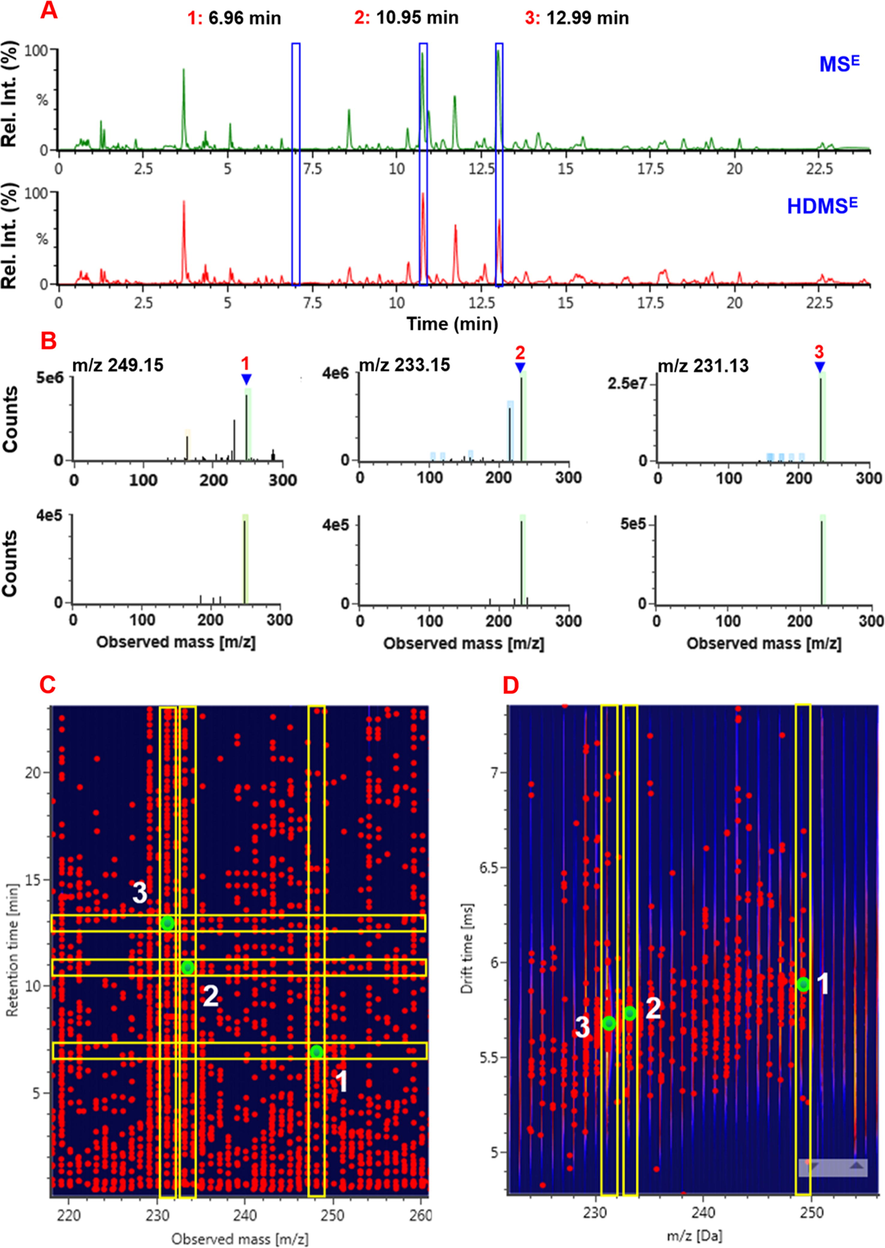
Comparison between the IM-enabled HDMSE and IM-disabled MSE in the positive ESI mode in profiling and charactering the multicomponents from Atractylodes macrocephala, using atractylenolides I (m/z 231.13), II (m/z 233.15), and III (m/z 249.15) as representatives. A-The base peak chromatograms recorded by MSE (up) and HDMSE (down); B-the full-scan MS1 spectra of atractylenolides I–III recorded by MSE (up) and HDMSE (down); C-the 3D plot recorded by MSE showing three peaks of atractylenolides I–III; D-2D-drift scope plot showing the additional separation of isomers by drift time.
3.4 Systematic characterization of the multicomponents from A. macrocephala based on the positive HDMSE data of eight fractionated samples by UNIFI-driven automatic peak annotation
Traditional methods used for the structural elucidation of TCM components based on the MSn data rely on manual interpretation, yielding the results closely correlated to the professional knowledge and largely different among the researchers. Therefore, we sought to establish highly efficient methods to analyze the MSn data that can generate reproducible characterization results. UNIFITM is a multifunctional bioinformatics platform enabling the automatic annotation of the high-resolution MS2 data obtained by MSE, DDA, HDMSE, and HDDDA, etc. (Radchenko et al., 2020; Zhang et al., 2019). Standardized workflows by applying UNIFI to annotate the positive HDMSE data were established in this work for the primary structural elucidation generating the lists of “Identified Components” and “Unknown Components”, while a confirming process based on the MS2 fragmentation pathways and retention characteristics was conducted which generated the final characterization list. The first-dimensional HILIC separation of the A. macrocephala extract yielded eight fractionated samples, and from Fig. 3 we were able to deduce that the multicomponents of A. macrocephala got enriched and numerous minor peaks were newly exposed. By analyzing the HDMSE data of these eight fractionated samples, as a result, we could identify or tentatively characterize a total of 251 components as given in Table S6, including 115 sesquiterpenoids, 90 polyacetylenes, 11 flavonoids, 9 benzoquinones, 12 coumarins, and 14 others. We emphasized and discussed the structural elucidation of the sesquiterpenoids, coumarins, flavonoids, and polyacetylenes, four subclasses of the major bioactive ingredients of A. macrocephala (Zhu et al., 2018).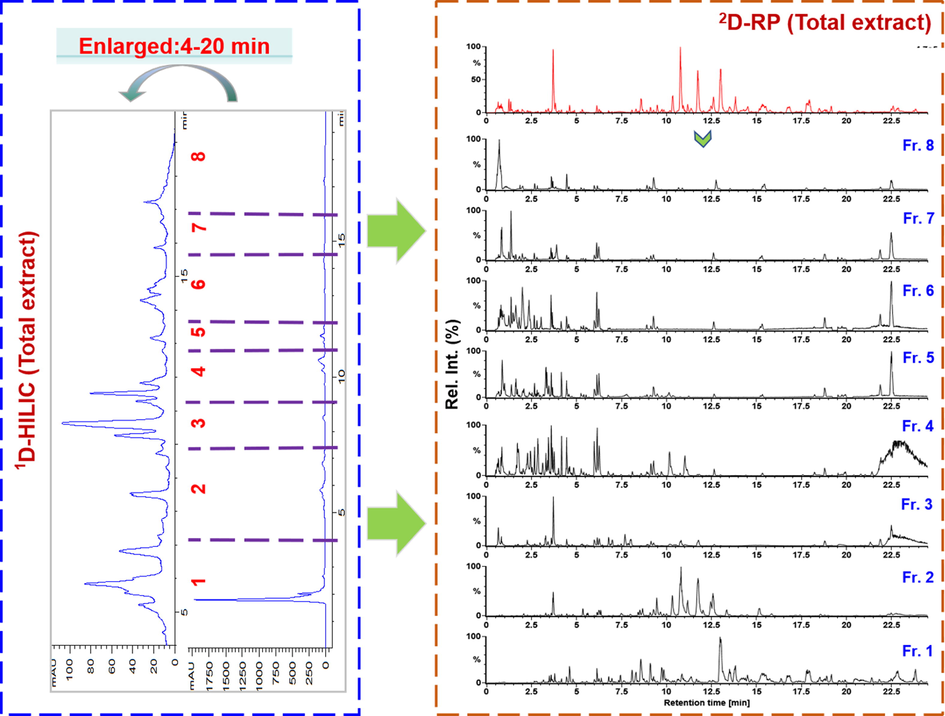
Illustration for the eluate collection by the first-dimensional HILIC separation of the total extract of Atractylodes macrocephala and the fractionation effect by comparing the positive-mode HDMSE spectra of the total extract and eight fractionated samples.
3.4.1 Sesquiterpenoids
Sesquiterpenoids are one of the most abundant and major bioactive constituents in A. macrocephala (Yang et al., 2016). The sesquiterpenoids (e.g. atractylenolides) have been extensively investigated and considered to have promising medicinal potential. Atractylenolides I–III are three representative compounds of this type, which are derived from atractylones by oxidative ring opening and dehydration. In this section, atractylenolide I (compound 190#, tR 12.99 min) was utilized as a reference to discuss the CID-MS2 fragmentation of sesquiterpenoids. In the MS1 spectrum, it gave the protonated precursor ion at m/z 231.1385, and the main cleavages occurred on rings A and C (Fig. 4). Fragmentation on the protonated molecule could open the lactone ring by eliminating H2O producing a fragment at m/z 213.1278, which was able to lose CO giving the product ion observed at m/z 185.1329. What’s more, the fragments of m/z 155.0859, 141.0703, 128.0624, and 115.0547 were obtained due to the additional cleavages of ring A on the ion m/z 185. The left section of ring B also generated the product ions at m/z 91.0543, 77.0387, and 65.0388, in the low-mass spectral region. The compound 248# (tR 22.37 min) produced a protonated precursor ion at m/z 463.2854 (C20H39O4+, mass error: 1.90 ppm), the CID-MS2 of which could produce a series of abundant product ions at m/z 447.2905, 231.1388, 185.1335, 163.0762, 143.0859, and 105.0705, which may be due to the eliminations of CH4, bimolecular fragmentation, and the loss of C2H4 after the breaking of ring C and ring A. These CID features were in accordance with the characteristic fragmentation pathways because of the lactone cleavage, and accordingly, we speculated compound 248# was dimerized by two atractylenolide II, tentatively characterized as biatractylenolide/biepiasterolide (Fig. 4) (Ma, Dou and Tian, 2020). In addition, compounds 105# (tR 6.96 min), 168# (tR 10.95 min), 191# (tR 13.00 min), and 227# (tR 18.04 min), were identified as atractylenolide III, atractylenolide II, atractylodin, and atractylon, respectively, as a result of the comparison with their retention time, precursor ions, and the fragmentation behavior with those of the reference compounds. Characterization of the other unknown sesquiterpenoids from A. macrocephala was mainly based on the positive CID-MS2 data analysis and searching against the established in-house library.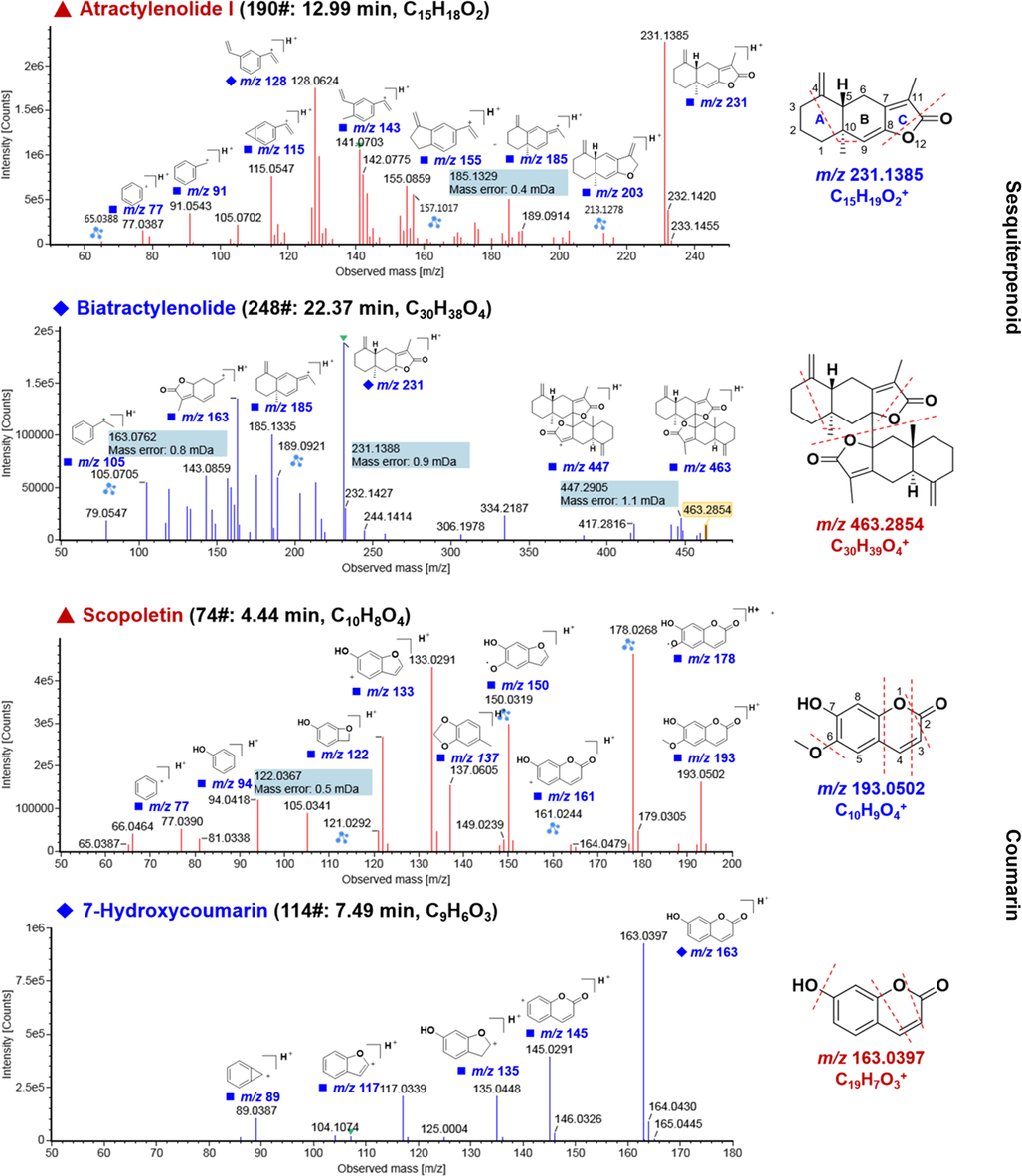
Illustration for the annotation of the positive CID-MS2 spectra of representative sesquiterpenoids (compounds 190# and 248#) and coumarins (compounds 74# and 114#).
3.4.2 Coumarins
Coumarins are a class of organic heterocyclic compounds with α-benzopyranone core structure, which widely exist in plants. The fragmentation behavior of coumarins was featured by the cleavages of the pyranone section by successive neutral loss of CO. Scopoletin (compound 74#, tR 4.44 min) was a simple coumarin which gave the protonated precursor ion at m/z 193.0502 (C10H9O4+, mass error: 3.20 ppm). Due to the presence of —OCH3, the fragments resulting from the easy elimination of free radical •CH3 and CH3OH were observed at m/z 178.0268 and 161.0244, respectively. The free radical fragment of m/z 178 could be further fragmented into the product ions by the successive losing of CO and 2 × CO giving the fragments at m/z 150.0319 and 122.0367, while the fragments at m/z 133.0291 and 105.0341 were dissociated from the ion of m/z 161 by the same fragmentation pathways. Moreover, CID-MS2 fragmentation of the precursor ion (m/z 193) could also directly eliminate two molecules of CO generating a product ion of m/z 137.0605. Accordingly, compound 74# was identified as scopoletin, which was confirmed by comparison with the reference compound (Fig. 4). Compound 114# (tR 7.49 min) was an unknown compound which gave a [M + H]+ precursor ion at m/z 163.0397 (C9H7O3+, mass error: 4.30 ppm), and accordingly the molecular formula was characterized as C9H6O3. In its CID-MS2 spectrum, some fragments, due to the continuous loss of CO, H2O, and CH3, were observed at m/z 135.0448 [M + H–CO]+, 117.0339 [M + H–CO–H2O]+, and 89.0387 [M + H–2CO–H2O]+, respectively. According to these evidences, compound 114# was tentatively characterized as 7-hydroxycoumarin or its isomer (Meng et al., 2019).
3.4.3 Flavonoids
Flavonoids have been known as the active ingredients of A. macrocephala with the antibacterial, anti-inflammatory, and other pharmacological activities (Zhu et al., 2018). Eleven compounds were identified as flavonoids from A. macrocephala, of which the characterization of the typical compounds 16# (tR 0.87 min), 64# (tR 3.88 min), 82# (tR 5.27 min), 88# (tR 5.56 min), and 97# (tR 6.29 min), were analyzed. Rich fragments were dissociated from compounds 88# (m/z 287.0556) and 97# (m/z 271.0610) at m/z 153.0194 [M + H–C8H6O2]+, 137.0608 [M + H–C8H5O3]+, 121.1018 [M + H–C8H5O4]+, 91.0544 [M + H–C9H7O5]+ and 165.0703 [M + H–C7H5O]+, 153.0701 [M + H–C8H6O]+, 105.0704 [M + H–C8H5O4]+, and 91.0546 [M + H–C9H7O4]+, respectively. These two compounds were identified as luteolin (88#) and apigenin (97#) by comparison with the authentic standards. The [M + H]+ precursors of an unknown compound 64# (m/z 449.1078) differed from that of compound 88# by 162 Da, which could be assigned as one molecule of glucose (Glc). Interestingly, the CID-MS2 of these two compounds underwent almost the same fragmentation pathways. Therefore, we could infer compound 64# was luteolin-O-glucoside. The unknown compounds 16# and 82# were more likely to be flavonones because of the characteristic product ions m/z 166.0867 and m/z 181.0500. Protonated precursor at m/z 451.1239 and m/z 449.1447, upon CID-MS2, could readily eliminate the attached Glc residue generating the aglycone ion at m/z 289.1406 and m/z 284.2952, respectively. By comparing the protonated precursors and the characteristic fragmentations, it could be inferred that the molecular formula difference between these two compounds was the CH3. Therefore, compound 16# was tentatively identified as scutellarein-O-glucoside, while compound 82# was characterized as hydroxy-methoxy-flavonone-O-glucoside.
3.4.4 Polyacetylenes
Polyacetylene is one class of characteristic components in A. macrocephala, for which the double bond is mainly of the trans-configuration. For polyacetylenes, CID-MS2 could produce multiple product ions by the neutral eliminations of CO, H2O, HCOOH, and CH3COOH, etc., together with the cleavages between the unsaturated olefinic and acetylenic bonds (Li et al., 2017). A total of 90 polyacetylene compounds were tentatively characterized, and most of them contained the 14-C skeleton of 2,8,10-trien-4,6-diyn-1,12,14-triol. Moreover, the hydroxyl group at C-12 /C-14 of the core structure was further esterified with acetic acid, 3-methylbut-2-enoic acid, and 3-methylbutanoic acid, etc.. In this study, we illustrated the characterization of four compounds (81#, 127#, 155#, and 189#) belonging to this structure subclass. In detail, the protonated ion of compound 81# (tR 4.87 min) was observed at m/z 233.1178, which, upon CID-MS2, yielded abundant product ions at m/z 215.1086, 187.0757, 152.0628, 141.0705, 128.0626, 115.0546, 105.0703, and 91.0544, respectively (Fig. 5), which were attributed to the loss of OH, C2H4O, C2H5O3, C3H7O3 C4H8O3, C5H9O3, C6H7O3, and C7H8O3, from the precursor ion. Similarly, in the case of compound 189# (tR 12.74 min), in addition to the protonated molecule at m/z 401.1971, characteristic ions at m/z 328.1673 ([M + H–C3H4O2]+), 313.1440 ([M + H–C4H7O2]+), 267.1024 ([M + H–C6H13O3]+), 205.0502 ([M + H–C12H19O2]+), 165.0707 ([M + H–C10H19O6]+) and 91.0546 ([C7H7]+), were observed. Based on the MS information, compounds 81# and 189# were tentatively characterized as 12-hydroxytetradeca-trien-4,6-diyn-1-ol (or its isomer) and 12-methylbutyryltetradeca-trien-4,6-diyne-1,14-diacetate (or its isomer), respectively (Chen, 1987; Yao and Yang, 2014). The protonated precursor ion of compound 155# (tR 9.97 min) at m/z 399.1603 was indicative of one molecule of H2 less than compound 189#. Very similar fragmentation pathways were observed for compound 155# producing diverse product ions of m/z 339.1603, 327.1588, 265.1236, 165.0703, and 91.0545. Therefore, compound 155# was tentatively characterized as 12-senecioyloxytetradeca-trien-4,6-diyne-1,14-diacetate (or its isomer). The MS/MS fragment ions of compound 127# (tR 8.34 min, [M + H]+ at m/z 359.1861) involved m/z 298.0890 ([M + H–C4H12]+), 267.1388 ([M + H–C5H11O2]+), 231.1385 ([M + H–C6H7O3]+), and 181.1019 ([C7H13O5]+). Based on the structure of compound 189# and the fragment information, it could be inferred that the acetyl group at C-1 of compound 127# should be replaced by OH. Therefore, we were able to putatively characterize compound 127# as 14-acetoxy-12-methylbutyryltetradeca-trien-4,6-diyn-1-ol (Yao and Yang, 2014).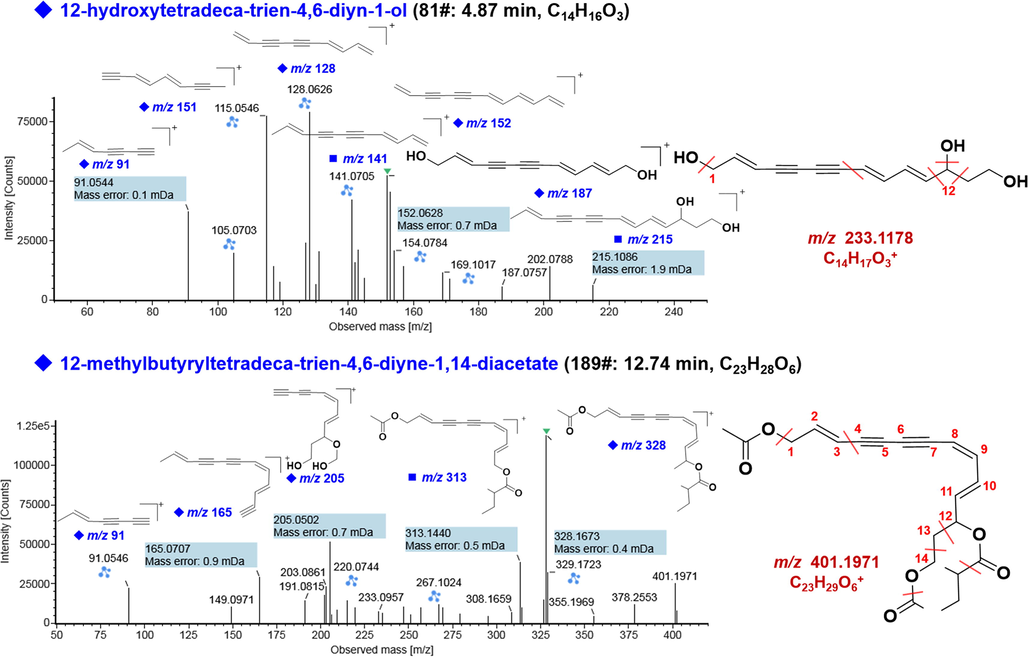
Illustration for the annotation of the positive CID-MS2 spectra of representative polyacetylenes (compounds 81# and 189#).
MS-based rapid chemical elucidation could primarily delineate the chemical composition of the medicinal plants. However, due to the lack of sufficient reference compounds, a major proportion of these compounds were only putatively characterized. We shall continue to perform the phytochemical research aiming to further validate the compounds identification results and support the bioactivity screening.
4 Conclusions
Targeting the complex multicomponents from a medicinal plant, A. macrocephalas, a dimension-enhanced four-dimensional separation approach, by offline 2D-LC coupled with IM-enabled HDMSE using a Vion IM-QTOF mass spectrometer, was successfully established and validated. High orthogonality (up to 0.91) was achieved due to the combination of an XBridge Amide column of HILIC and a BEH C18AX column of mixed IEC and RPC modes. The application of data-independent HDMSE followed by UNIFI-driven automatic peak annotation greatly improved the efficiency, reliability, and reproducibility in structural elucidation of the plant metabolites with rich fragmentation information provided. The enabling of IM separation could significantly boost the resolution of ions in both MS1 and MS2 spectra. Beneficial to these merits, we could identify or tentatively characterize a total of 251 components (including 115 sesquiterpenoids, 90 polyacetylenes, 11 flavonoids, 9 benzoquinones, 12 coumarins, and 14 others) from A. macrocephalas, which indicated great improvement in profiling and characterizing the multicomponents, in contrast to the previous reports by conventional LC/MS. The integral offline 2D-LC/IM-QTOF-HDMSE approach can facilitate the four-dimensional separation, particularly applicable to the in-depth characterization of the multicomponents from medicinal plants. The results obtained in this work can further benefit the bioactive compounds discovery, quality control, and pharmacological researches of Atractylodis Macrocephalae Rhizoma, as the continuous efforts.
Acknowledgments
Funding: This research was funded by National Key Research and Development Program of China (Grant No. 2018YFC1704500, 2018YFC1707904, 2018YFC1707905, and 2017YFC1702104), National Science & Technology Major Project of China (Grant No. 2018ZX09201011, 2018ZX09735-002, and 2018ZX09711001-009-010), and National Natural Science Foundation of China (Grant No. 81872996).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- A new measure of orthogonality for multidimensional chromatography. Anal. Chim. Acta.. 2014;838:93-101.
- [Google Scholar]
- Comprehensively qualitative and quantitative analysis of ginsenosides in Panax notoginseng leaves by online two-dimensional liquid chromatography coupled to hybrid linear ion trap Orbitrap mass spectrometry with deeply optimized dilution and modulation system. Anal. Chim. Acta.. 2019;1079:237-251.
- [Google Scholar]
- Adding a new separation dimension to MS and LC-MS: What is the utility of ion mobility spectrometry. J. Sep. Sci.. 2018;41:20-67.
- [Google Scholar]
- Anti-inflammatory components isolated from Atractylodes macrocephala Koidz. Nat. Prod. Res.. 2008;22:1418-1427.
- [Google Scholar]
- Simultaneously targeted and untargeted multicomponent characterization of Erzhi Pill by offline two-dimensional liquid chromatography/quadrupole-Orbitrap mass spectrometry. J. Chromatogr. A. 2018;1584:87-96.
- [Google Scholar]
- Recent advances on HPLC/MS in medicinal plant analysis-An update covering 2011–2016. J. Pharm. Biomed. Anal.. 2018;147:211-233.
- [Google Scholar]
- Simultaneous profiling and holistic comparison of the metabolomes among the flower buds of Panax ginseng, Panax quinquefolius, and Panax notoginseng by UHPLC/IM-QTOF-HDMSE-based metabolomics analysis. Molecules. 2019;24:2188.
- [Google Scholar]
- Recent development in liquid chromatography stationary phases for separation of Traditional Chinese Medicine components. J. Pharmaceut. Biomed. Anal.. 2016;130:336-346.
- [CrossRef] [Google Scholar]
- Anti-cancer effect of Atractylodes macrocephala extract by double induction of apoptotic and autophagic cell death in head and neck cancer cells. Bangladesh J. Pharmacol.. 2017;12:140-146.
- [Google Scholar]
- High resolution LC-MS characterization of phenolic compounds and the evaluation of antioxidant properties of a tropical purple radish genotype. Arab. J. Chem.. 2020;13:1355-1366.
- [Google Scholar]
- Identification and mass spectrum fragmentation study of polyacetylenes constituents from the rhizomes of Atractylodis macrocephalae and Atractylodes lancea. Chin. J. New drugs. 2017;26(9):1071-1078.
- [Google Scholar]
- Study on comprehensive quality evaluation of Atractylodes macrocephala based on constituents and pharmacodynamics. Med. Plant. 2014;5:30-32.
- [Google Scholar]
- Integration of multicomponent characterization, untargeted metabolomics and mass spectrometry imaging to unveil the holistic chemical transformations and key markers associated with wine steaming of Ligustri Lucidi Fructus. J. Chromatogr. A.. 2020;1624:461228
- [Google Scholar]
- A modified data filtering strategy for targeted characterization of polymers in complex matrixes using drift tube ion mobility-mass spectrometry: Application to analysis of procyanidins in the grape seed extracts. Food Chem.. 2020;321:126693
- [Google Scholar]
- Enhanced identification of the in vivo metabolites of Ecliptae Herba in rat plasma by integrating untargeted data-dependent MS2 and predictive multiple reaction monitoring-information dependent acquisition-enhanced product ion scan. J. Chromatogr. B. 2019;1109:99-111.
- [Google Scholar]
- Natural disesquiterpenoids: an overview of their chemical structures, pharmacological activities, and biosynthetic pathways. Phytochem. Rev. 2020:1-61.
- [Google Scholar]
- Isomer detection on the basis of analyte adduct formation with the components of the mobile phase and tandem mass spectrometry. Arab. J. Chem.. 2019;12:181-187.
- [Google Scholar]
- Analysis of the composition of raw and processed Atractylodes macrocephala Koidz by UPLC/Q-TOF-MS. China Food Addit.. 2019;4:145-153.
- [Google Scholar]
- Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal. Chem.. 2015;87:1137-1144.
- [Google Scholar]
- Hydrophilic interaction chromatography (HILIC) for the determination of cetirizine dihydrochloride. Arab. J. Chem.. 2019;12:4204-4211.
- [Google Scholar]
- An enhanced strategy integrating offline two-dimensional separation and step-wise precursor ion list-based raster-mass defect filter: Characterization of indole alkaloids in five botanical origins of Uncariae Ramulus Cum Unicis as an exemplary application. J. Chromatogr. A. 2018;1563:392-397.
- [Google Scholar]
- An integrated approach for global profiling of multi-type constituents: Comprehensive chemical characterization of Lonicerae Japonicae Flos as a case study. J. Chromatogr. A. 2020;1613:460674
- [Google Scholar]
- Recent developments in two-dimensional liquid chromatography: fundamental improvements for practical applications. Anal. Chem.. 2019;91:250-263.
- [Google Scholar]
- Global profiling and novel structure discovery using multiple neutral loss/precursor ion scanning combined with substructure recognition and statistical analysis (MNPSS): Characterization of terpene-conjugated curcuminoids in Curcuma longa as a case study. Anal. Chem.. 2016;88:703-710.
- [Google Scholar]
- A green protocol for efficient discovery of novel natural compounds: characterization of new ginsenosides from the stem and leaves of Panax ginseng. Anal. Chim. Acta. 2015;893:65-76.
- [Google Scholar]
- Metabolite identification using ion mobility enhanced data-independent acquisition strategy and automated data processing. Rapid Commun. Mass Spectrom.. 2020;34:e8792
- [Google Scholar]
- Liquid chromatography coupled to mass spectrometry for metabolite profiling in the field of drug discovery. Expert Opin. Drug Discov.. 2019;14:469-483.
- [Google Scholar]
- Cross-mixing study of a poisonous Cestrum species, Cestrum diurnum in herbal raw material by chemical fingerprinting using LC-ESI-QTOF-MS/MS. Arab. J. Chem.. 2020;13:7851-7859.
- [Google Scholar]
- Metabolomic study of raw and processed Atractylodes macrocephala Koidz by LC-MS. J. Pharm. Biomed. Anal.. 2014;98:74-84.
- [Google Scholar]
- Simultaneous determination of atractylenolide II and atractylenolide III by liquid chromatography-tandem mass spectrometry in rat plasma and its application in a pharmacokinetic study after oral administration of Atractylodes Macrocephala Rhizoma extract. Biomed. Chromatogr.. 2012;26:1386-1392.
- [Google Scholar]
- A classification of liquid chromatography mass spectrometry techniques for evaluation of chemical composition and quality control of traditional medicines. J. Chromatogr. A. 2020;1609:460501
- [Google Scholar]
- Influence of sulfur fumigation on the chemical profiles of Atractylodes macrocephala Koidz. evaluated by UFLC–QTOF–MS combined with multivariate statistical analysis. J. Pharm. Biomed. Anal.. 2017;141:19-31.
- [Google Scholar]
- The emerging of ion mobility-mass spectrometry in lipidomics to facilitate lipid separation and identification. Trends Anal. Chem.. 2019;116:332-339.
- [Google Scholar]
- Holistic quality evaluation of Saposhnikoviae Radix (Saposhnikovia divaricata) by reversed-phase ultra-high performance liquid chromatography and hydrophilic interaction chromatography coupled with ion mobility quadrupole time-of-flight mass spectrometry-based untargeted metabolomics. Arab. J. Chem.. 2020;13:8835-8847.
- [Google Scholar]
- Rapid screening and characterization of triterpene saponins in Acanthopanax senticosus leaves via untargeted MSAll and SWATH techniques on a quadrupole time of flight mass spectrometry. J. Pharm. Biomed. Anal.. 2019;170:68-82.
- [Google Scholar]
- The effects of supplementing diets with Atractylodes macrocephala Koidz rhizomes on growth performance and immune function in piglets. J. Anim. Feed Sci.. 2012;21:302-312.
- [Google Scholar]
- A generic multiple reaction monitoring based approach for plant flavonoids profiling using a triple quadrupole linear ion trap mass spectrometry. J. Am. Soc. Mass Spectrom.. 2014;25:955-965.
- [Google Scholar]
- Method development and application of offline two-dimensional liquid chromatography/quadrupole time-of-flight mass spectrometry-fast data directed analysis for comprehensive characterization of the saponins from Xueshuantong Injection. J. Pharm. Biomed. Anal.. 2016;128:322-332.
- [Google Scholar]
- Approaches to establish Q-markers for the quality standards of traditional Chinese medicine. Acta. Pharm. Sin. B.. 2017;7:439-446.
- [Google Scholar]
- Global profiling combined with predicted metabolites screening for discovery of natural compounds: characterization of ginsenosides in the leaves of Panax notoginseng ad a case study. J. Chromatogr. A. 2018;1538:34-44.
- [Google Scholar]
- Bioactivity-guided isolation of polyacetylenes with inhibitory activity against NO production in LPS-activated RAW264.7 macrophages from the rhizomes of Atractylodes macrocephala. J. Ethnopharmacol.. 2014;151:791-799.
- [Google Scholar]
- DecoMetDIA: deconvolution of multiplexed MS/MS spectra for metabolite identification in SWATH-MS based untargeted metabolomics. Anal. Chem.. 2019;91:11897-11904.
- [Google Scholar]
- Integration of data-dependent acquisition (DDA) and data-independent high-definition MSE (HDMSE) for the comprehensive profiling and characterization of multicomponents from Panax japonicus by UHPLC/IM-QTOF-MS. Molecules. 2019;24:2708.
- [Google Scholar]
- A multidimensional analytical approach based on time-decoupled online comprehensive two-dimensional liquid chromatography coupled with ion mobility quadrupole time-of-flight mass spectrometry for the analysis of ginsenosides from white and red ginsengs. J. Pharm. Biomed. Anal.. 2018;163:24-33.
- [Google Scholar]
- Application of two-dimensional liquid chromatography in the separation of traditional Chinese medicine. J. Sep. Sci.. 2020;43:87-104.
- [Google Scholar]
- Large-scale prediction of collision cross-section values for metabolites in ion mobility-mass spectrometry. Anal. Chem.. 2016;88:11084-11091.
- [Google Scholar]
- LipidCCS: prediction of collision cross-section values for lipids with high precision to support ion mobility-mass spectrometry-based lipidomics. Anal. Chem.. 2017;89:9559-9566.
- [Google Scholar]
- The traditional uses, phytochemistry, and pharmacology of Atractylodes macrocephala Koidz.: A review. J. Ethnopharmacol.. 2018;226:143-167.
- [Google Scholar]
- Data-dependent acquisition and database-driven efficient peak annotation for the comprehensive profiling and characterization of the multicomponents from compound Xueshuantong capsule by UHPLC/IM-QTOF-MS. Molecule. 2019;24:3431.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2020.102957.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







