Translate this page into:
A novel inhibitor, 2-cyano-3-(1-phenyl-3-(thiophen-2-yl)-pyrazol-4-yl)acrylamide linked to sulphamethoxazole, blocks anti-apoptotic proteins via molecular docking and strongly induced apoptosis of HCT116 cell line by different molecular tools
⁎Corresponding authors. magdafikry85@yahoo.com (Magda F. Mohamed), ismail_shafy@yahoo.com (Ismail A. Abdelhamid)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Abstract
A new series of cyanoacrylamides incorporating sulphamethoxazol were prepared and confirmed by different spectral tools. Anticancer screening of the new compounds was done against three different types of carcinoma cell lines involving (A549, HCT116, and MDA) using MTT assay. Compound 7 among all tested derivatives achieved the best cytotoxic effect against all tested carcinoma cell lines. HCT116 revealed the best sensitivity and cytotoxic activity toward compound 7 relative to 5-FU. The target compound offered less toxic effect when tested on normal melanocytes (HFB4). Simulation modeling studies revealed strong binding affinity toward the following domains (1dls, 2c6o, and 2wgj) and moderate binding modes toward (3eyl, 4kmp, 2w3l, and 5lab) domains with different binding energy scores. Gene expression profile outlined that caspase-3, BAX, and P53 genes were strongly upregulated relative to their control, while BCL2, MMP1, and CDK2 were effectively down regulated assuming the activation of the apoptotic pathway. Flow cytometry technique revealed that compound 7 stimulated cell cycle arrests at the G2/M phase. Other extensive molecular diagnostic tools were utilized in this report as ELISA, DPA, SEM, and TEM assays which confirmed that our target novel compound 7 was a very promising and interesting chemotherapeutic agent with less toxic effect. Also, authors herein suggested that additional sulphamethoxazole linked to 3-(1-phenyl-3-(thiophen-2-yl)-1H-pyrazol-4-yl)acrylonitrile in compound 7 was responsible for its promising cytotoxic activity against colorectal carcinoma cell line.
Keywords
Colorectal carcinoma
Cyanoacrylamides
Sulphamethoxazole
Cytotoxicity
Modeling studies
Apoptosis
1 Introduction
Millions of people continue to fall sick with cancer disease each year all over the word especially developed countries. So, our purpose and challenges in this article was to find new chemotherapeutic agent that can be used as a cure for cancer and less toxic in the same time. Compounds incorporation acrylamide group have pronounced biological applications as antifungal (Bartkiene et al., 2018; El-Gaby et al., 2002b; Ren et al., 2018), antimicrobial (El-Gaby et al., 2002a; El-Gaby et al., 2000; Malviya et al., 2019; Nasr et al., 2020), antidiabetic (Asif et al., 2019; Maren, 1976; Dogo et al., 2019) anticancer (Fadda et al., 2012; Jiao et al., 2019; Mohamed et al., 2014; Mubarak and Al-Hamdani, 2019; Zhang et al., 2019) and anti-inflammatory properties (Roifman et al., 2000). In particular, it has been reported that 3-aryl-2-cyanoacrylamide scaffold incorporating sulfonamide moieties was used as inhibitors of the metallo-enzyme carbonic anhydrase and against the cytosolic human (h) isoforms hCA I and II, as well as the trans-membrane, tumor associated ones CA IX and XII, which are validated antitumor targets (Alafeefy et al., 2013; Del Prete et al., 2020). Moreover, the great interest in the sulphamethoxazole has been stimulated by some promising pharmaceutical activities such as anticancer (Ghorab et al., 2009; Gupta et al., 2013) and antimicrobial agents (Eldesouky et al., 2018; Hida et al., 2005; Tondolo et al., 2018; Underwood et al., 2011; Zander et al., 2010; Zhanel et al., 2000). In addition, it can be used in the treatment of urinary tract infections (McCarty et al., 1999; Raz et al., 2002; Zhanel et al., 2000). Thus, it is assumed that combining the above mentioned acrylamide and sulphamethoxazole into a molecular framework may afford molecules with high biological activity. Based on the above assumptions and in a continuation to our interest directed towards the synthesis of bioactive heterocyclic compounds (Ghozlan et al., 2015; Mohamed et al., 2014, 2012). We report herein the synthesis of some new derivatives of cyanoacrylamides incorporated with sulphamethoxazole moiety and test their cytotoxic effect by MTT assay against different cancer cell lines. We also report the molecular docking and molecular tools studies to suggest the concrete action of the prepared drugs both theoretically and experimentally.
2 Materials and method
2.1 Chemistry
Melting points were determined on a Stuart melting point apparatus and are uncorrected. The IR spectra were measured as KBr pellets on a FTIR Bruker-Vector 22 spectrophotometer. 1H and 13C NMR spectra were measured using Bruker Ultrashield 400 MHz or Ascend 400 MHz (1H: 400 MHz, 13C: 100.6 MHz) instruments using TMS as internal standard. Mass spectra were measured on a Shimadzu GCMS-Q-1000 EX mass spectrometer at 70 eV. The elemental analyses were carried out at the Micro-Analytical Center, Cairo University using Automated analyzer CHNS, Vario EL III, Elementar, Germany.
2.1.1 2-Cyano-N-(4-{[(5-methylisoxazol-3-yl)amino]sulfonyl}phenyl)acetamide (3)
Sulphamethaxazole(10 mmol) 1 was added to 3-(3,5-dimethyl-1H-pyrazol-1-yl)-3-oxopropanenitrile 2 (10 mmol) in dry toluene (20 mL). The mixture was heated at reflux for 3 h. The solvent was evaporated and crude product was purified by crystallization from ethanol to give white crystals (90%). Mp 216–218 °C. IR (KBr): νmax/cm−1 3331, 3279 (2NH), 2264 (CN), 1690 (CO). 1H NMR (400 MHz, DMSO‑d6): δ 2.30 (s, 3H, CH3), 3.96 (s, 2H, CH2), 6.13 (s, 1H, isoxazole-CH), 7.73–7.75 (d, 2H, Ar-H, J = 8.8 Hz), 7.82–7.85 (d, 2H, Ar-H, J = 8.8 Hz,), 10.71 (s, 1H, NH), 11.37 (br, 1H, NH) ppm. 13C NMR (100 MHz, DMSO‑d6): δ 12.5, 27.5, 95.9, 116.1, 119.6, 128.7, 134.4, 143.1, 158.0, 162.5, 170.9 ppm. MS (EI, 70 eV): 320 [M+]. Anal. Calcd. for C13H12N4O4S: C, 48.75; H, 3.78; N, 17.49. Found C, 48.46; H, 3.53; N, 17.28.
2.1.2 General procedure for synthesis of compounds 5–9
A mixture of compound 3 (1 mmol) and appropriate aldehyde 4a-e (1 mmol) was heated at reflux in absolute EtOH (20 mL) containing piperidine (0.2 mL, 2 mmol) for 60 min. The crude product was collected and crystallized from EtOH.
2.1.3 2-Cyano-3-(1,3-diphenyl-1H-pyrazol-4-yl)-N-(4-(N-(5-methylisoxazol-3-yl)sulfamoyl)phenyl)acrylamide (5)
Pale yellow crystals (89%). Mp: 264–266 °C. IR (KBr): νmax/cm−1 3315, 3210 (2 NH), 2215 (CN), 1635 (CO). 1H NMR (400 MHz, DMSO‑d6): δ 2.29 (s, 3H, CH3), 6.13 (s, 1H, CH), 7.44–7.96 (m, 14H, Ar-H), 8.18 (s, 1H, vinyl-H), 9.22 (s, 1H, pyrazole-H), 10.69 (s, 1H, NH), 11.35 (s, 1H, NH). 13C NMR (DMSO‑d6): δ 12.5, 95.9, 105.1, 114.8, 117.0, 120.1, 121.0, 128.3, 128.5, 129.4, 129.5, 129.8, 129.9, 130.4, 131.3, 134.7, 139.0, 143.0, 143.1, 155.2, 158.1, 161.3, 170.7. MS (EI, 70 eV): 550 [M+]. Anal. Calcd. for C29H22N6O4S: C, 63.26; H, 4.03; N, 15.26. Found: C, 63.48; H, 4.22; N, 15.41.
2.1.4 2-Cyano-3-(3-(4-hydroxyphenyl)-1-phenyl-1H-pyrazol-4-yl)-N-(4-(N-(5-methylisoxazol-3-yl)sulfamoyl)phenyl)acrylamide (6)
Pale yellow crystals (82%). Mp: 232–234 °C. IR (KBr): νmax/cm−1 3321, 3195 (2 NH), 2219 (CN), 1676 (CO). 1H NMR (400 MHz, DMSO‑d6): δ 2.16 (s, 3H, CH3), 5.85 (s, 1H, CH), 6.93 (d, 2H, Ar-H, J = 8.4 Hz), 7.44–7.70 (m, 10H, Ar-H and OH), 7.91 (d, 2H, Ar-H, J = 8.4 Hz), 8.14 (s, 1H, vinyl-H), 9.16 (s, 1H, pyrazole-H), 10.55 (br. s, 2H, 2 NH). 13C NMR (DMSO‑d6): δ 12.7, 96.9, 104.7, 114.6, 116.3, 117.2, 119.9, 120.6, 121.9, 127.4, 128.3, 129.6, 130.3, 130.8, 139.1, 140.5, 141.2, 143.1, 155.4, 159.0, 161.0, 164.5, 167.5.MS (EI, 70 eV): 566 [M+]. Anal. Calcd. for C29H22N6O5S: C, 61.48; H, 3.91; N, 14.83. Found: C, 61.63; H, 4.11; N, 14.96.
2.1.5 2-Cyano-N-(4-(N-(5-methylisoxazol-3-yl)sulfamoyl)phenyl)-3-(1-phenyl-3-(thiophen-2-yl)-1H-pyrazol-4-yl)acrylamide (7)
Pale yellow crystals (86%). Mp: 260–262 °C. IR (KBr): νmax/cm−1 3315, 3115 (2 NH), 2215 (CN), 1638 (CO). 1H NMR (400 MHz, DMSO‑d6): δ 2.30 (s, 3H, CH3), 6.13 (s, 1H, CH), 7.25–7.94 (m, 12H, Ar-H and thiophene-H), 8.33 (s, 1H, vinyl-H), 9.21 (s, 1H, pyrazole-H), 10.72 (s, 1H, NH), 11.35 (s, 1H, NH). 13C NMR (DMSO‑d6): δ 12.7, 97.1, 106.0, 114.5, 117.0, 120.0, 120.4, 127.2, 128.5, 128.7, 128.9, 129.0, 130.1, 130.4, 132.7, 138.8, 139.8, 141.8, 142.8, 149.0, 160.8, 165.9, 166.8.MS (EI, 70 eV): 556 [M+]. Anal. Calcd. for C27H20N6O4S2: C, 58.26; H, 3.62; N, 15.10. Found: C, 58.38; H, 3.83; N, 15.19.
2.1.6 2-Cyano-N-(4-(N-(5-methylisoxazol-3-yl)sulfamoyl)phenyl)-3-(thiophen-2-yl)acrylamide (8)
Pale yellow crystals (84%). Mp: 283–285 °C. IR (KBr): νmax/cm−1 3310, 3112 (2 NH), 2215 (CN), 1677 (CO). 1H NMR (400 MHz, DMSO‑d6): δ 2.30 (s, 3H, CH3), 6.13 (s, 1H, CH), 7.33–8.16 (m, 7H, Ar-H), 8.54 (s, 1H, vinyl-H, 10.62 (s, 1H, NH), 11.35 (s, 1H, NH). MS (EI, 70 eV): 414 [M+]. Anal. Calcd. for C18H14N4O4S2: C, 52.16; H, 3.40; N, 13.52. Found: C, 52.31; H, 3.44; N, 13.38.
2.1.7 2-Cyano-3-(furan-2-yl)-N-(4-(N-(5-methylisoxazol-3-yl)sulfamoyl)phenyl)acrylamide (9)
Pale yellow crystals (86%). Mp: 253–255 °C. IR (KBr): νmax/cm−1 3321, 3112 (2 NH), 2215 (CN), 1676 (CO).. 1H NMR (400 MHz, DMSO‑d6): δ 2.29 (s, 3H, CH3), 6.11 (s, 1H, CH), 6.86–6.87 (m, 1H, Furan-H), 7.44 (d, 1H, furan-H, J = 3.6 Hz), 7.84 (s, 4H, Ar-H), 8.18 (s, 1H, vinyl-H), 8.19, (d, 1H, Furan-H, J = 3.6 Hz), 10.58 (s, 1H, NH), 11.35 (s, 1H, NH). 13C NMR (DMSO‑d6): 12.5, 95.9, 101.8, 114.6, 116.3, 120.8, 122.9, 128.3, 134.2, 135.1, 137.1, 142.9, 148.6, 158.5, 161.4, 170.5. MS (EI, 70 eV): 398 [M+]. Anal. Calcd. for C18H14N4O5S: C, 54.27; H, 3.54; N, 14.06. Found: C, 54.34; H, 3.68; N, 14.28.
2.2 Bioactivity
2.2.1 MTT assay
Cytotoxic evaluations of the newly synthesized cyanoacrylamide incorporating sulphamethoxazol derivatives (3, 5–9) were evaluated to different carcinoma cell lines as A549 (lung carcinoma cell line), HCT116 (Colorectal carcinoma cell line), and MDA (breast carcinoma cell line). In addition, toxicity test was performed on the most active compound 7 against HFB4 (Normal human skin melanocytes cell line) for 48 h post treatment using MTT assay. All cell lines were purchased from American Tissue Culture
Collection (Rockville, MD, USA), and cultured at 37 °C with 5% carbonic acid gas in an exceedingly humidified atmosphere in RPMI-1640 media supplemented with 10% fetal bovine serum. In order to examine the possibility of novel structures, 5-FU, a commercial classical anticancer drug was used as a reference organic drug. All compounds and 5-FU have been dissolved in DMSO to prepare stock concentrations of 5 mg/ml. The stock solutions were filtered via 0.22 lm syringe filter. Double fold dilutions were prepared through including equal volumes of the dissolved compounds to fresh RPMI-1640 culture medium. Ninety-six-well cancer cells pre-cultured plates were treated with descending double fold serially diluted compounds and 5-FU at 37 °C for 24 h. Negative cell culture control was involved. Residual living cells were treated with 50 µl of filtered MTT (5 mg/mL) at 37 °C for 4 h. MTT was discarded and plates were PBS washed three times. DMSO was added as 50 µl for each well. Plates were shaken on a plate shaker for 30 min to dissolve the produced complex of intracellular blue formazan. Optical densities (O. Ds) were measured at 570 nm using an ELISA plate reader. Viability percentages were calculated and the IC50 values of all compounds and 5-FU were determined by the prism program. Informational data were stated for three independent experiments and existing as mean ± S.D.
2.2.2 Molecular modelling
The newly designed compound 7 was applied to the docking program, using (MOE) program 2009.10 version to predict the binding mode between the ligand (compound 7) and the active domains of the randomly selected proteins for theoretical studies (Mohamed et al., 2018). The construction was subjected to energy diminishing via Hamiltonian-Force Field-MMFF94x and Gasteiger partial charges have been delivered to ligand atoms. Non-polar hydrogen atoms were merged, and rotatable bonds had been defined. Eventually, the compound structure was saved in the docking software in each the 2D and the 3D formulas which were prepared for docking. Protein version changed into obtained from Brookhaven Protein information financial institution (www.RCSB.org). The crystal structure of the active sites of tested proteins (2wgj, 1dls, 2c6o, 3eyl, 4kmp, 2w3l, 5lab, and 4wt2) that was complexed with the following standard ligands (sorafenib, methotrexate, triazolopyrimidine, Smac, hydroxypyrrolidine derivative, Phenyl Tetrahydroisoquinoline Amide Complex, NNGH, and AM-7209 respectively, were downloaded. Then treated programmatically to be utilized in predicting the binding fashions of the novel designed compound 7 to the selected active domains. Proteins became prepared for docking as follows: (i) all standard ligands cocrystalized with proteins became eliminated; (ii) The specific binding domains, chain C, turned into selected; (iii) The grid container dimensions were selected to be sufficiently huge to consist of all surrounding crucial residues; (iv) Proteins were 3D protonated, where hydrogen atoms have been brought at their widespread geometry, the partial expenses were computed and the system turned into optimized. Then, the protein became geared up for docking. All docking calculations had been done on the co-crystallized ligand and target ligand 7 with different protein versions. All remaining steps were followed according to our literatures (Mohamed et al., 2018), and finally, data was visualized using BIOVIA Discovery Studio V6.1.0.15350 program.
2.2.3 Real time polymerase chain reaction (qPCR)
Total RNA was isolated from control sample and compound 7 (42.9 µg/ml) treated HCT116 cells and then purified, using a kit that was provided by Qiagen RNA extraction, according to the manufacturer’s protocol. The yield of the entire RNA gained was assayed at 260 nm spectrophotometrically. The quantitative real time polymerase chain reaction was done subsequently by supplies that had been provided by Bioline, a median life science company, UK (SensiFAST™ SYBR® Hi-ROX One-Phase Kit, Catalog No. PI- 50217 V). This kit had been formulated for highly reproducible first-strand cDNA synthesis and subsequent real-time PCR in a single tube. The ideal primer pair was selected to include the considered factors as melting temperature (Tm: 60–65 °C), GC content (40–60%) and amplicon length of about 90–200 bp. Primer sequences (sense and anti-sense) for the target six genes (p53, caspase3, bcl2, bax, MMP1, and CDK2) and the reference housekeeping gene (GAPDH) were illustrated in (Table 1). RT-PCR master mix was prepared according to the followed protocol, then 4 µl RNA template was added to get a reaction mixture of 20 µl final volume. The prepared reaction mix samples were applied in real time PCR (Step One Applied Bio-system, Foster city, USA). Sensi FAST™ SYBR® Hi-ROX One-Step Kit is compatible with three-step cycling as follows: One cycle at 45 °C for 10 min allowing reverse transcription, one cycle at 95 °C for 2 min allowing polymerase activation and 40 cycles at 95 °C, 60 °C, and 72 °C for 5, 10 and 5 s allowing denaturation, annealing, and extension respectively (acquire at the end of step). After the RT-PCR run, the data were expressed in Cycle threshold (Ct). The PCR data sheet comprises Ct values of evaluated genes (p53, caspase3, bcl2, bax, MMP1, and CDK2) and the reference housekeeping gene (GAPDH). The relative quantitation (RQ) of each target gene is quantified according to the calculation of delta-delta Ct (ΔΔCt). Data were reported for three independent experiments. We calculated the RQ of each gene by taking 2−ΔΔCt as following:
ΔΔCt = [(Ct target; Sample)_(Ct ref; Sample)]_[(Ct target; Control)_Ct ref; Control)]
| Genes | Primer Sequences |
|---|---|
| MMP1 | F-5′-CTGGCCACAACTGCCAAATG-3′ |
| R-5′-CTGTCCCTGAACAGTACTTA-3′. | |
| CDK2 | F-5′-TCAGAAGCCATCTCTTCTGTG-3′ |
| R-5′-TACATCTGGAGGCCAGTCAAT-3′ | |
| Caspase3 | F-5′-TTC ATT ATT CAG GCC TGC CGA GG-3′ |
| R-5′-TTC TGA CAG GCC ATG TCA TCC TCA-3′ | |
| P53 | F-5′- CCCCTCCTGGCCCCTGTCATCTTC-3′ |
| R-5′-GCAGCGCCTCACAACCTCCGTCAT-3′ | |
| Bax | F-5′-GTTTCA TCC AGG ATC GAG CAG-3′ |
| R-5′-CATCTT CTT CCA GAT GGT GA-3′ | |
| Bcl2 | F-5′-CCTGTG GAT GAC TGA GTA CC-3′ |
| R-5′-GAGACA GCC AGG AGA AAT CA-3′ | |
| GAPDH | F-5′-TTCCAGGACCAAGATCCCTCCAAA-3′ |
| F-5′-TTCCAGGACCAAGATCCCTCCAAA-3′ | |
2.2.4 DNA fragmentation assay
DNA fragmentation steps were performed using the diphenylamine method (DPA) for colorimetric assay according to the previously described literature (Zhao et al., 2013). This assay involved the generation of two types of DNA by centrifugation step, intact one and fragmented species after lysis of HCT116 cells treated with (42.9 µg/ml) of compound 7. Then precipitation and quantitation by (DPA) reagent. All procedures and details were performed according to the literature (Stamos et al., 2002), where 1.0 mL of cells suspension (not less than 5 × 105 and no more than 5 × 106) were utilized, in order to obtain an OD600 for DNA > 0.04 and <1.200) in tubes labeled B (bottom). Cells were centrifuged at 200g at 4 °C for 10 min, then supernatants were carefully transferred in new tubes labeled S (supernatant). The pellet in tubes B were vortexed vigorously in 1.0 mL TTE solution, and this step is allowable to release of fragmented chromatin from nuclei, once the cell lysis (due to the occurrence of Triton X-100 in the TTE solution) and disturbance of the nuclear construction (following Mg++ chelation by EDTA in the TTE solution). Tubes B were centrifuged to separate fragmented DNA from intact chromatin, centrifuge at 20,000g for 10 min at 4 °C, then supernatants were transferred carefully in new tubes labeled T. All remaining instructions were completed according to the above literature where the percentage of fragmented DNA can be calculated using the formula:
%Fragmented DNA = S + T × 100
S + T + B
2.2.5 Morphological characterization
-
Scanning electron microscopy (SEM)
To prepare the cells for morphological and ultrastructural analysis, they were cultivated in RPMI-1640 media for 24 then treated with 42.9 µg/ml of compound 7 for 48 h of incubation at 37 °C with 5% CO2 in a humidified atmosphere. Then cells were collected and washed several times with PBS and kept in glutaraldehydes. The samples were evaluated by scanning electron microscopy (SEM), following instructions previously described in the literature (Ishiwata et al., 2017) with minor modifications 5% gluteraldehyde was used for cell fixation in phosphate buffer (pH 7.4) at 4 °C overnight. Afterwards, the samples were dehydrated with a graded ethanol series (30–50-70–90-100% for 30 min each) followed by hexamethyldisilazane (HDMS) for 10 min. The membranes were then cut with a scalpel, put on a carbon disc and sputtered with gold before they were examined with the SEM. Images were taken with a Jeol microscope (JSM-639OLA, Japan) under high pressure conditions with a secondary electron detector and using 10 kV acceleration voltage.
Transmission electron microscopy
TEM imaging used to be accomplished in TEM lab FA-CURP, Faculty of Agriculture, Cairo University Research Park. After incubation period about 48 h,each control and treated HCT116 cells with 42.9 µg/ml of compound 7 for 48 h had been harvested by the use of trypsin and centrifuged for 10 min at 1000 rpm and room temperature. Cell pellets have been processed for TEM via fixation in PBS containing 2.5% glutaraldehyde. Samples have been postfixed in 1% osmium tetroxide, dehydrated in graded alcohol and embedded in an epoxy resin. Microtome sections organized at approximately 500–1000 lm thickness with a Leica Ultra cut UCT extremely microtome. Thin sections had been stained with toluidine blue (1x) then sections have been examined using camera Lica ICC50HD. Ultra-thin sections organized at about 75–90 µm thickness and were stained with uranyl acetate and lead citrate, then examined via transmission electron microscope JEOL, (JoelL td., Tokyo, Japan), (JEM-1400 TEM) at the candidate magnification. Then images have been collected by AMT model of CCD camera.
2.2.6 Cell cycle analysis
The procedures of this assay were performed in more detail according to our literature (Ali et al., 2017). Cells (3 × 106) were seeded in two culture flasks for manipulating and test for 48 h, then, cells had been treated with the IC50concentration of the examined new compound 7. After incubation for 48 h, single cell suspensions have been prepared by means of treating the cells with a trypsin/EDTA combination in RPMI. Spontaneously detached cells present in culture medium have been additionally covered in the analysis. Cell Suspensions (1 × 106) had been pelleted, re-suspended in 200 µl of phosphate buffered saline (PBS) and fixed for at least 30 min at 4 °C in 2 mL of 70% ice-cold ethanol. Cells were washed two times in PBS, re-suspended in 800 µl PBS to which was added 100 of 1 mg/ml RNase A and 100 µl of 400 µg/ml propidium iodide (PI), and allowed to incubate for 30 min at 37 °C. Cells were analysed by Epics XL-MCL flow cytometer (Beckman Coulter, Miami, FL). The distribution of cell cycle had been analysed by Multi Cycle software (Flow Systems of Phoenix, San Diego, CA) and various phases of cell cycle occupied by different percentage of cells, namely G0/G1, S, and G2/M, was calculated. Three independent experiments were done and the data presented are the average values.
2.2.7 Annexin V apoptosis assay
Apoptosis evaluation was carried out the using annexin V–FITC Kit Apoptosis Detection Kit. Seed cells (3 × 106 cells) in 4 culture-flasks, two for control and two for taking a look at for 48 h. Cells in check-flasks were treated with 42.9 µg/ml of the newly examined compound 7 for 48 h. After incubation period, single cell suspensions from control and treated cells were organized through treating the cells with a trypsin/EDTA combination in RPMI. Spontaneously detached cells existing in culture medium have been also included in the analysis. Cell suspensions (1 × 106) were pelleted, washed with PBS and centrifuged for 5 min at 500g at 4 °C. Supernatant was discarded and the cell pellets have been re-suspended in ice-cold (1x) binding buffer to 5 × 106 cells/ml, tubes had been stored on ice exactly 5 µl of dissolved PI and 1 µl annexin V-FITC solution had been added to 100 µl of the cell suspensions and combined quietly. Tubes were kept on the ice and incubated for 15 min in the dark. 400 µl of ice-cold (1x) binding buffer was one time introduced and combined gently. Cell preparations have been analyzed within 30 min through going with the flow cytometry. The test was once repeated three instances and the data presented were the average values.
3 Results and discussions
3.1 Synthetic chemistry
The starting 2-cyano-N-(4-(N-(5-methylisoxazol-3-yl)sulfamoyl)phenyl)acetamide 3 was obtained in high yields and purity via the direct cyanoacetylation of sulphamethoxazole1 with 1-cyanoacetyl-3,5-dimethylpyrazole 2 following the reported procedures (Scheme 1) (Gorobets et al., 2004; Mohamed et al., 2017a, 2017b; Štetinová et al., 1996). Knoevenagel condensation reaction of the cyanoacetylsulphamethoxazole3 with the mole equivalent of heteroaldehydes (3-aryl-1-phenyl-1H-pyrazole-4-carbaldehyde 4a-c, thiophene2-carbaldehyde 4d or furane-2-carbaldehyde 4e) in the presence of basic catalyst as piperidine affords the corresponding cyanoacrylamides incorporating sulphamethoxazole 5–9 which many have Z or E configuration. Although the 1H and 13C NMR cannot simply differentiate between the two isomers, our structures were assumed to have E configuration based on the X-Ray crystal structure of similar compounds in literature (Dyachenko et al., 2007). The structures of the title compounds were confirmed by inspection of their spectral data. For example, the mass spectrum of compound 7 revealed molecular ion peak as a base peak at m/z 556. 1H NMR displayed a singlet signal at δ 2.30 ppm for the methyl attached to isoxazole ring. Also, it indicated two singlet signals at δ 6.13 and 8.33 for the isoxazole-H4 and vinyl-H3 protons respectively. Moreover, the amide and sulphonamide NH groups appeared as two broad signal at δ 10.72 and 11.35 ppm. The signals of aromatic protons appear as multiplets in the area of 7.25–7.94 ppm. Furthermore, the 13C NMR (APT) indicated 23 signals related to 23 different carbons. It featured the characteristic methyl, cyano and carbonyl groups at δ 12.7, 117, and 166.8 ppm respectively. All other carbons appear at their expected positions.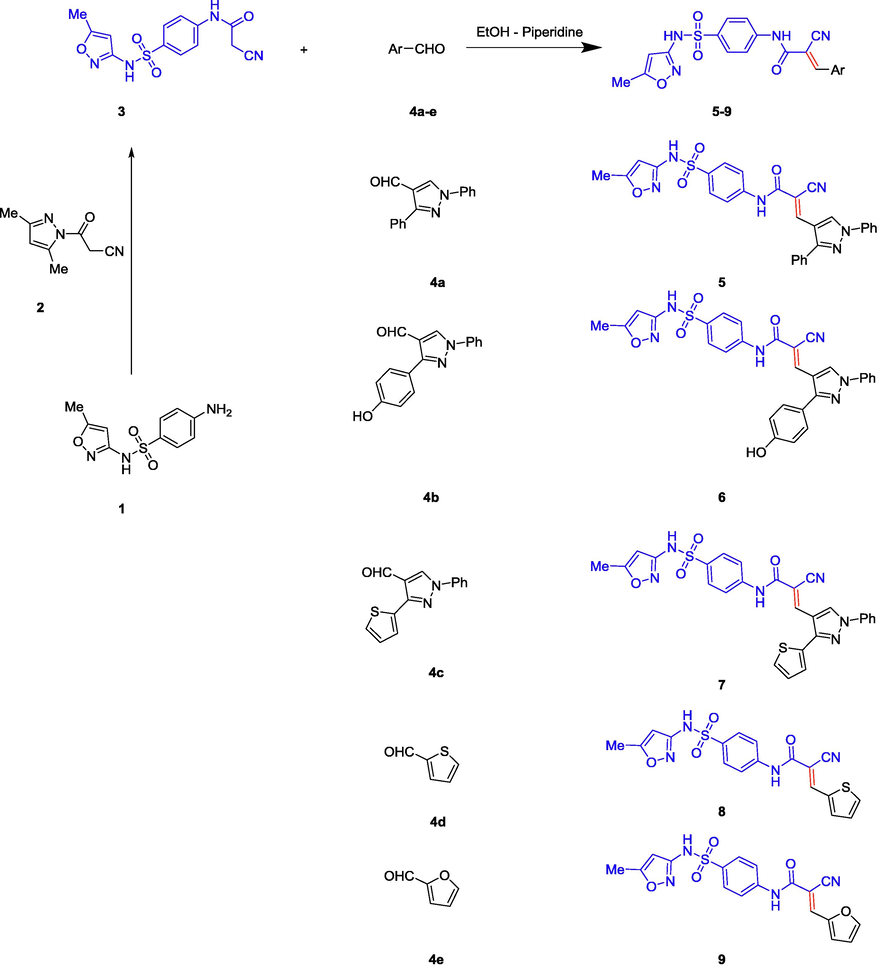
Synthesis of cyanoacrylamides incorporating sulphamethoxazole 5–9.
3.2 Bioactivity
3.2.1 Cytotoxicity
Cytotoxic assay was performed on a new series of very interesting biologically active cyanoacrylamide incorporating sulphamethoxazole derivatives (5–9) which presented in Scheme 1. All measurements illustrate that all novels (5–9) exhibit very strong and promising sensitivity towards all tested cancer cell lines relative to our lead compound 3 (Table 2). Generally, colon carcinoma revealed potent sensitivity toward all synthesized derivatives including the lead compound. Also, breast carcinoma gets the best sensitivity to our derivatives especially compound 5 which achieved the lowest IC50 value (34.6 µg/ml) compared to the lead compound 3 and the standard control 5-fluorouracil (5-FU) with IC50 (581.1, 205.2 µg/ml) respectively. The cyanoacrylamide incorporating both sulphamethoxazole and 3-(thiophen-2-yl)-1H-pyrazole group 7 represents the most active and promising cytotoxic agent against the selected cell lines with IC50 values (61.3, 47.5, and 42.9 µg/ml) respectively, this meant that the additional 1-phenyl-3-(thiophen-2-yl)-4-vinylpyrazole group into the lead compound 3 withIC50 (828.3, 581.1, and 498.4 µg/ml), strongly enhanced the cytotoxic effect of compound 7 as outlined in Fig. 1 (cf. SI). Regarding lung carcinoma, the new addition of 4-vinylpyrazole as in compounds 5 and 6 promoted and enhanced IC50 values to become (115.8, and 77.3 µg/ml) respectively, relative to their lead compound 3. In addition, they were considered more promising than their positive control value (284.2 µg/ml). The additional 2-vinylthiophene and 2-vinylfuran groups into compounds 8 and 9 respectively, strongly induced their activity with respect to the lead compound 3 but, on the other hand they showed moderate activity regarding the positive control (5-FU). With respect to MDA carcinoma, the addition of 4-vinylpyrazole group into compound 5, and 7 were strongly promoted where they offered highly sensitive action toward this line with lowest inhibition concentration (34.6, and 47.5 µg/ml) as depicted in Table 2 and Fig. 1 (cf. SI). Moreover, compound 9 exposed promising activity toward breast carcinoma when compared to lead compound 3 and 5-FU, while showed a moderate effect regarding lung and colon carcinoma taking into consideration readings of the standard control (5-FU). In case of colon carcinoma, derivatives 6 and 7 illustrated the highest activity with IC50 values (87.1, 42.9 µg/ml) respectively, following them compound 5 which also showed strong effect toward this line. In addition, compound 8 still presented high cytotoxic effect relative to the positive control, but unfortunately compound 9 was not promising toward this line when compared to both lead compound 3 and the positive control (5-FU). However, before a new chemotherapeutic candidate could enter in the clinical trials, it has to be evaluated for its safety. The new derivative 7 was tested for its safety on human normal melanocytes (HFB4), it exhibited less toxic effect toward this normal line with inhibition concentration (1750 µg/ml) as outlined in Fig. 2 (cf. SI). This large reading IC50 value confirmed that it was perfect and selective anticancer agent. In this new prepared series, our novel 2-cyanoacrylamides achieved more promising and potent effect rather than series published by us in the previous literatures (Mohamed et al., 2017a, 2017b), and this means that the additional sulfamethoxazole group greatly enhances the cytotoxic effect and biological activities, especially compound 7 which considered the best sensitive and selective anticancer agent toward all tested carcinoma. Furthermore, in attempts to find out the mechanism of inhibition of compound 7 towards colon carcinoma, theoretical (simulation) and molecular works were performed.
Compounds
IC50 (µg/ml)
A549
MDA
HCT116
HFB4
Lead compound 3
828.32
581.1
498.4
–
Compound 5
115.8
34.6
169.7
–
Compound 6
77.39
122.2
87.1
–
Compound 7*
61.32
47.5
42.9
1750
Compound 8
387.7
319.9
346.1
–
Compound 9
315.9
149.6
427.8
–
5FU
284.2
205.22
500.11
–
3.2.2 Modeling studies
The molecular modeling method, including ranking and computational screening of ligand libraries to identify the potential lead chemical candidates. A lot of studies using simulating programs have shown that these scannings have a higher enrichment of active drugs than random screening tools (Mohamed et al., 2018). In this new interesting study, we wish to suggest the molecular action of our active designed derivative 7 through different theoretical studies to provide possibilities, effort and save time. We used two different docking program softwares to illustrate the inhibitory effect of the newly tested inhibitor into the active site of some antiapoptotic proteins. The current report is revolved on screening and testing different binding modes of component 7 which was the most potent compound selected from the training set to different domains of protein markers. The crystal structures of the tested active domains (Anaplastic Lymphoma Kinase (Alk), dihydrofolate reductase enzyme (DHFR), human CDK2, xIAP-BIR3, human bcl2-xl, mmp12, and mdm2 (PDB codes: 2wgj, 1dls, 2c6o, 3eyl, 4kmp, 2w3l, 5lab, and 4wt2) were in a complex formula with co-crystallized ligands (sorafenib, methotrexate, triazolopyrimidine, Smac, hydroxypyrrolidine derivative, tetrahydroisoquinoline amide complex, NNGH, and AM-7209) respectively, as a reference inhibitor ligands (Cossu et al., 2009; Cui et al., 2011; Lewis et al., 1995; Richardson et al., 2006; Stamos et al., 2002) were utilized to check the binding affinities of the seven tested proteins bound to our novel compound 7 within the molecular operating program (MOE). Fig. 3 depicted that 2D and 3D dimensional models of all selected active domains with compound 7, where data illustrated the most suitable binding energy scores obtained from simulating compound 7 to different binding sites (2wgj, 1dls, 2c6o, 3eyl, 4kmp, 2w3l, 5lab, and 4wt2) as follow (−31.345, −21.415, −25.312, −20.498, −21.562, −22.222, −18.921, and −23.881 Kcal/mol) respectively, relative to energy scores of standard co-crystallized ligands (−32.197, −34.770, −24.936, −24.976, −24.191, −18.219, −15.201, and −44.431 Kcal/mol) respectively. The strongest bindings achieved in this study were illustrated by fitting compound 7 within the active domains of dihydrofolate reductase enzyme (DHFR) and human CDK2 domain (2c6o) respectively. Compound 7 was rounded tightly by different amino acid sequences of 1dls and 2c6o domains respectively. Also, higher activity was recorded by Anaplastic Lymphoma Kinase (Alk), which fitted strongly to compound 7 through different residues. In addition, moderate binding was recorded against (3eyl, 4kmp, 2w3l, and 5lab) domains. Moreover, less binding affinity herein was achieved with MDM2 (4wt2) domain. The different types of interactions in Fig. 3 were represented by the following colored dot lines, conventional hydrogen bond (green lines), carbon hydrogen bonds (faint green lines), Pi-Pi stacked interaction (dark violet line), Pi-alkyl interactions (faint purple lines), Pi-cation interactions (faint brown lines), Pi-anion (deep brown lines), and vander-waals contact (green residues). The highest number of interactions were recorded with the binding of compound 7 with dihydrofolate reductase enzyme (Fig. 3ii). Compound 7 fitted into the active domain of enzyme through very strong four hydrogen bonds with different amino acids residue. Other types of interactions were recorded with the same enzyme as Pi-anion interactions. Also, promising binding affinity achieved by incorporating compound 7 into the human CDK2 active domain (Fig. 3iii) through different interactions, three conventional hydrogen bonds, two carbon - hydrogen bonds, one Pi-cation, two Pi-anion, and four Pi-alkyl interactions. In addition, our novel achieved three arene-cation interactions with anaplastic lymphoma kinase (2wgj) domain. On the other hand, a moderate binding mode was exerted by the action of compound 7 on the following apoptotic inhibitions domains (3eyl, 4kmp, 2w3l). Regarding proteins responsible for inhibitions of apoptosis xIAP-BIR3, the first domain illustrated one hydrogen bond and one arene-cation interaction with compound 7 through the same amino acid as illustrated in (Fig. 3iv). The other two domains exerted only one arene-cation affinity toward our target compound with the different amino acid residue (Fig. 3v and vi) respectively. The moderate binding was also achieved regarding the mdm2 domain (Fig. 3vii) into the tested compound. Unfortunately, the weak binding affinity of compound 7 was showed against mdm2 (4wt2) as illustrated in (Fig. 3viii). Generally, in this theoretical studies all molecular docking studies outlined that compound 7 bounded to all domains by vander-waals contact through different residues. It was noted from modeling results in Fig. 3 that the cyanoacrylamide group enhanced the biological activity binding affinity of this novel toward different domains and this was compatible to our previous literature (Mohamed et al., 2017a). Also, the new addition of sulfamethoxazole group in this recent report improved strongly the inhibition or blocking the tested domains. In addition to (SO2) group increased the blocking mode of the domains (2wgj, 1dls, 2c6o, 2w3l, 5lab, 4wt2 and 3eyl) and hence enhanced the biological activity of the target molecule 7. Different binding affinity modes obtained from this study illustrated that the designed compound 7 showed promising binding affinity toward most of proteins set selected from many trials and thus initiated apoptosis of different cancer types.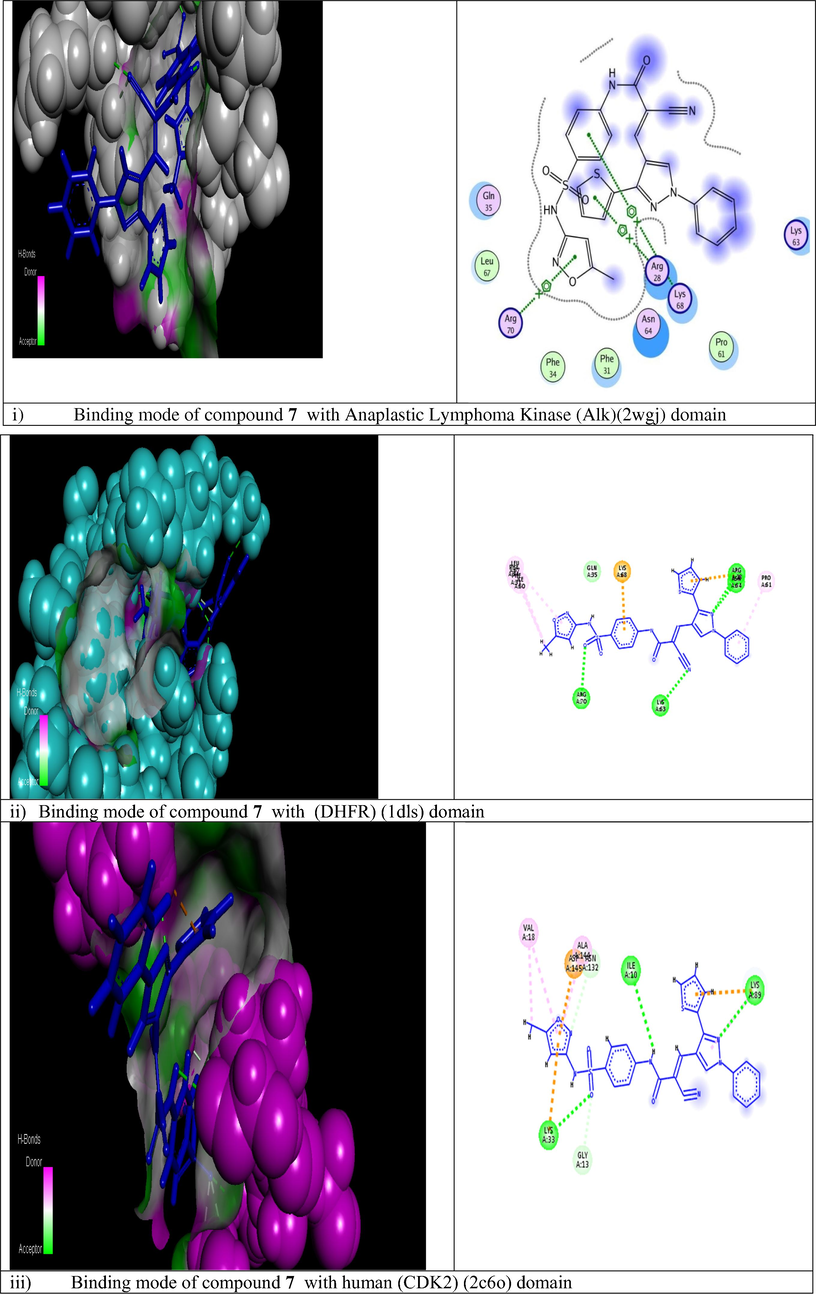
Modeling representations in 3D and 2D dimensional modes of compound 7 with the following active domains (2wgj, 1dls, 2c6o, 3eyl, 4kmp, 2w3l, 5lab, and 4wt2) respectively.
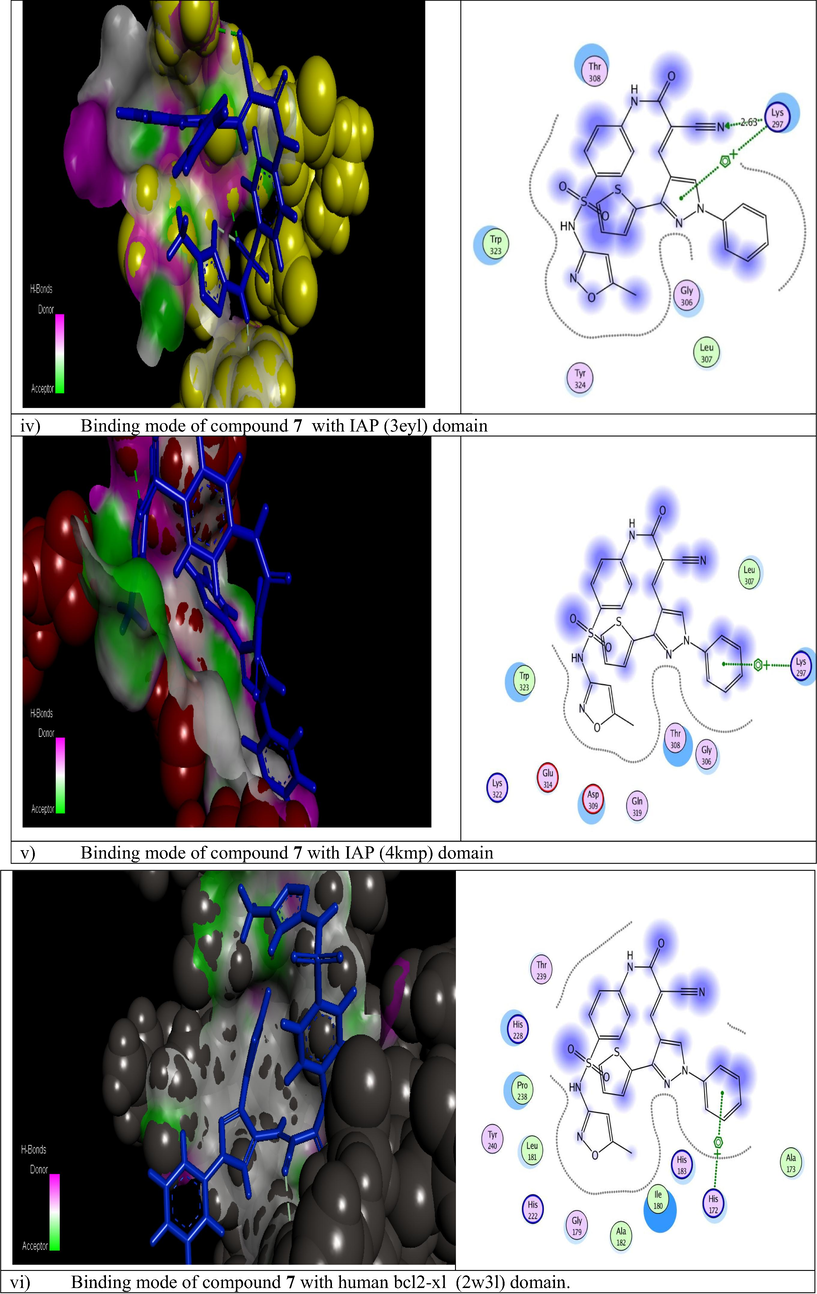
Modeling representations in 3D and 2D dimensional modes of compound 7 with the following active domains (2wgj, 1dls, 2c6o, 3eyl, 4kmp, 2w3l, 5lab, and 4wt2) respectively.
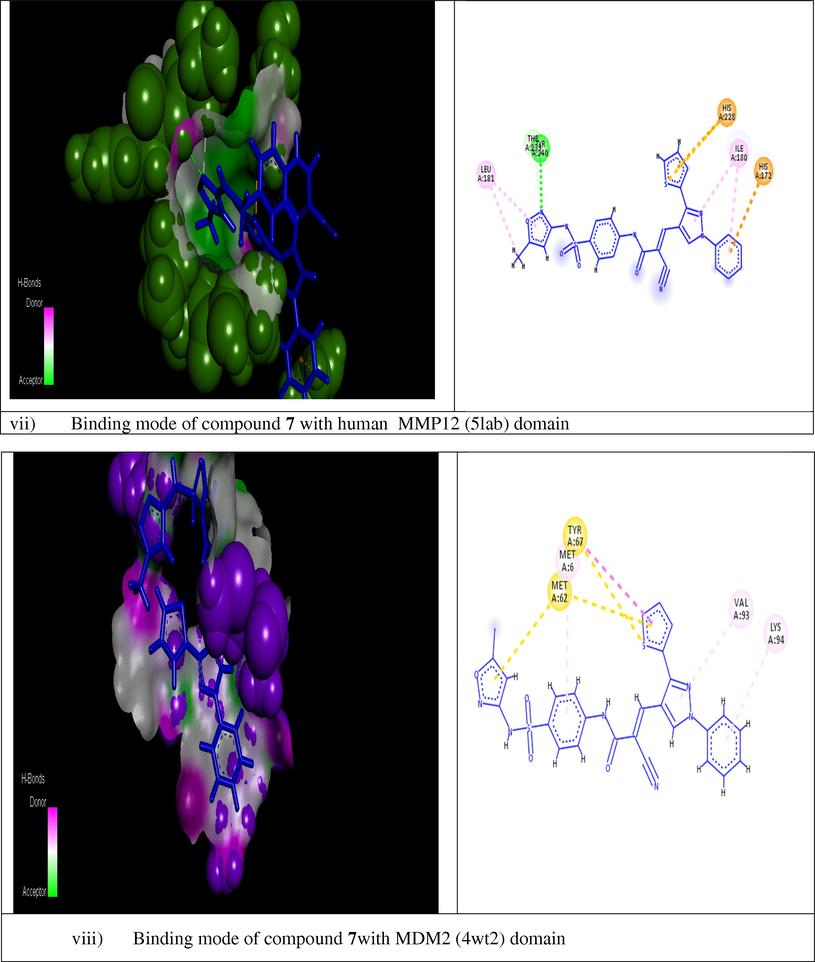
Modeling representations in 3D and 2D dimensional modes of compound 7 with the following active domains (2wgj, 1dls, 2c6o, 3eyl, 4kmp, 2w3l, 5lab, and 4wt2) respectively.
3.2.3 Real time polymerase chain reaction
The novel compound 7 was selected from this new series for molecular studies on one of the three sensitive cell lines (colon carcinoma) as it illustrated high sensitivity and cytotoxicity toward the tested cell line as mentioned in the previous section. It exhibited the lowest IC50 value (42.9 µg/ml) (as shown in Table 2), and this novel was considered less toxic toward human normal melanocytes (HfB4). Analysis of the relative expression of genes that controlling apoptotic process was important and argent step in determining the mechanism that our novel follow inside the cancer cell, so HCT116 cells were treated with IC50 value of the compound 7 for 48 h at 37 °C, and then cells for both control and sample were collected for real-time PCR analysis of the following genes (BAX, P53, Caspase-3, BCL2, MMP1, and CDK2), using specific primers for each one. Fig. 4 represented that compounds 7 strongly stimulated the expression of pro-apoptotic BAX gene with (fold change = 6.383), and apoptotic Caspase-3 gene with (fold change = 7.875), In addition, the expression of the tumor suppressor gene p53 was enhanced strongly by the action of the new compound 7 (fold change = 16.226). On the other side, compound 7 significantly depressed the expression level of anti-apoptotic genes CDK2 (fold change = 0.559), MMP1 (fold change = 0.244), and BCL2 (fold change = 0.143). Thus, increased the induction levels of BAX, P53, and caspase-3, and down regulation of CDK2, MMP1, and BCL2 genes by novel compound 7 relative to control proposed the growth inhibition and induction of apoptotic pathway in the treated colon carcinoma.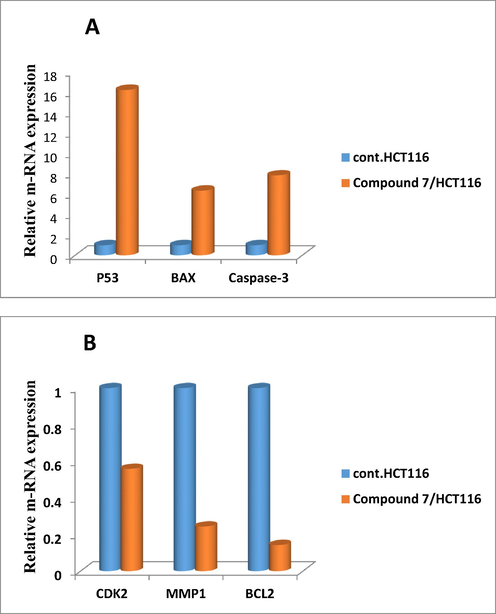
Relative gene expression representation of apoptotic proteins A (BAX, P53, and Caspase-3) and anti-apoptotic protein B (BCL2, MMP1, and CDK2) for colon cancer cell lines treated with compound 7, versus control using real time PCR tool.
3.2.4 The effect of compound 7 on genomic DNA fragmentation of colorectal carcinoma
Failure to repair DNA due to severe damage may result in transcriptional and replication blockage, mutagenesis, and/or cell cytotoxicity (Alarcon-Vargas and Ronai, 2002). This has been shown to be involved in a variety of genetically inherited disorders, in aging (Matsuoka et al., 2000) and in carcinogenesis (Maya et al., 2001; Shieh et al., 2000). This lead to initiation of apoptosis. Herein, colorimetrical quantization diphenylamine (DPA) method was utilized to determine the percentage of fragmented DNA in genomic DNA sample of colon cell lines treated with IC50 value (42.9 µg/ml) of compound 7. Fig. 5 represents a schematic diagram that showed the percentage of fragmented DNA in both colon cancer cells treated with compound 7 and untreated sample (control). This data exposed that novel compound 7 was strong and more effective in damaging the genomic DNA of HCT116 cells. Results showed distinctly a high percentage of damage caused by compound 7 (34.73282%) relative to the percent of fragmentation in control sample (6.953224%), thus this data confirmed high cytotoxic and selective action of compound 7. Generally, it has been suggested that degradation of the macromolecule in cells undergoing apoptosis was initially limited to internucleosomal DNA fragmentation. A last report (Kaufmann et al., 1993), however, outlined that internucleosomal DNA degradation in human leukemia cells treated with etoposide was accompanied by enzymatic quantitative degradation of (pADPRp) to stable fragments. An analogous fragmentation of pADPRp was showed in Molt4 cells treated with the inhibitor (5-deazaacyclotetrahydrofolate). In distinction to this consequence, earlier studies designated that human K562 cells, a cell type previously described to resist diphtheria toxin-prompted apoptosis (Chang et al., 1989), failed to show evidence of endonucleolytic DNA degradation and pADPRp cleavage. These observations strengthen the association between endonucleolytic DNA degradation and pADPRp fragmentation. It was unclear that whether this pADPRp cleavage was limited to certain cell lines treated with certain agents or whether it was a general phenomenon taking place in a wide variety of cell lines undergoing chemotherapeutic agents-induced apoptosis.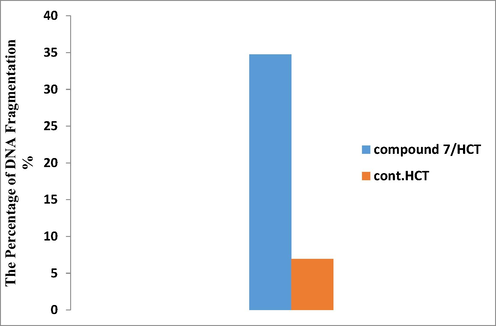
Schematic diagram represented colorimetric DPA assay which indicated the percent of DNA fragmentation of colon carcinoma treated with compound 7 relative to control.
3.2.5 Apoptotic protein markers assay for (caspase-3, cytochrome-c, and VEGFR) by ELISA
Human VEGFR, Cytochrome c, and Caspase-3 concentrations were intended for precise quantitative measurement by Human in vitro (Enzyme-Linked Immunosorbent Assay) ELISA kits for the lysate of cell. Apoptotic cell death is a fundamental feature of virtually all cells. The highly coordinated manner of this induced cell death suggested that the cells activated a common death program, towards which diverse signal-transducing pathways converge. The mitochondria turned out to participate in the main control or executioner phase of the cell death cascade. Cytochrome c was recognized as a component essential for the critical steps in apoptosis, caspase-3 activation, and DNA fragmentation. Cytochrome-c was exhibited to redistribute from mitochondria to cytosol during apoptosis in integral cells. Measurement of Cytochrome c released from the mitochondria is a tool to identify the first initial phases for initiating apoptosis in cells. With respect to caspases, they were essential in the regulation of apoptotic process. Many cancer cell lines were found expressing caspase-3 protein, with highest expression level in immune cells. Caspase-3 was synthesized as inactive proenzyme, where upon cleavage at Asp175/Ser176, was converted to the active form. The data obtained revealed that our new derivative 7 was a strong promising derivative which induced a significant increase in protein levels of caspase-3 and cytochrome-c as compared to their relative negative control samples at its IC50 value of 42.9 µg/ml (Table 2). The schematic diagram showed in (Fig. 6A and B) indicated that compound 7 induced significant increase in both caspase-3 and cytochrome-c proteins activity (450.9, and 0.9976 pg/ml, respectively) relative to their controls (17.27, and 0.03053 pg/ml) respectively. Angiogenesis, was an essential process for tumor growth and development. A process which induced by tumor hypoxia and resulted in several and complexed angiogenic factor profile proceeded as a result of genomic DNA instability in the tumor cells. So, It wasn’t surprising that targeted neutralization of a single angiogenic factor, has been the focus for anti-angiogenic cancer therapy (Block et al., 2015). VEGF significantly affect the vascular permeability and was potent angiogenic factor in many bioassays. It played an important role in the developmental stages supporting the proliferation of blood vessels. In this assay, it has been illustrated that inhibition of VEGF protein activity by treatment with a monoclonal antibody specific for VEGF can suppress tumor growth. Also, data presented in Table 2 reflected the potency of our novel compound 7 in down regulated vascular endothelial growth factors (VEGF) expressed by the tumor cells for surviving and metastasis to lesser extent (586.7 Pg/ml) compared to its control sample (1487). In conclusion, compound 7 was a very promising chemotherapeutic agent which significantly unregulated the expression level of caspase-3 (the promoter of apoptosis), and cytochrome-c proteins. In addition, it was strongly down regulated the anti-apoptotic protein (VEGF) (Fig. 6A) (see Table 3).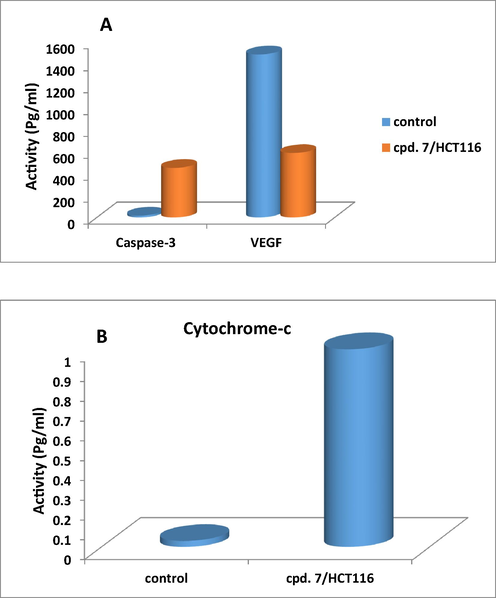
Schematic diagram represented the effect of compound 7 on the expression level of Caspase 3, Cytochrome-c, and VEGFR proteins in colorectal cell line (HCT116) for 48 h of treatment.
Caspase-3
Cytochrome-c
VEGF
Samples
Pg/ml
FLD
Pg/ml
FLD
Pg/ml
FLD
Cont.HCT116
17.27 ± 1.2a
1
0.03053 ± 0.01a
1
586.7 ± 19.4a
0.394
Compound 7/HCT116
450.9 ± 17b
26.10886
0.9976 ± 0.03b
32.67606
1487 ± 39.61b
1
Data are presented as Mean ± SD. Means values with different marked superscript letters were significant (P < 0.05).
3.2.6 Microscopic examination of colorectal cells treated with compound 7
-
Scanning electron microscopy (SEM)
In our recent study, to investigate the effects of compound 7 on HCT116 cells at the ultrastructural pattern, different changes have to be examined on the surface topography of HCT116 cells that can lead to cell apoptosis in response to this new treatment using SEM. Scanning electron microscopy (SEM) has a unique and powerful abilities in medical, biophysical, and biological research. It provided novel information about organelles, membrane structure, and the cytoskeleton of cells. SEM was a distinctive tool in identifying and quantifying the morphological features of apoptosis. To prepare the cells for morphological and ultrastructural analysis, they were cultivated in RPMI-1640 media for 24 then treated with 42.9 µg/ml of compound 7 for 48 h of incubation at 37 °C with 5% CO2 in a humidified atmosphere. The samples were evaluated by scanning electron microscopy (SEM), following the manuscript instructions described in the experimental section. It was very interesting technique to check the integrity of the cell membrane structure of colorectal carcinoma through comparing the difference in structures before and after treatment with the tested compound, where the cell morphology has great effect on different biological processes. SEM was a none destructive tool which used to obtain a surface image of cells in three-dimension phase and gives us important features about the status of the cell membrane. In this analysis, SEM was used for cell surface scanning and to observe a variety of changes in surface morphology. Cell surface topography of cells treated with compound 7 changed tremendously relative to the negative control. Also, the cell surface ultra-structure in test, and control presented obvious differences. Regarding the control, none treated HCT116 cells (Fig. 7A), a relatively uniform or homogeneous surface was illustrated. Fig. 7B depicted many small membrane protrusions and larger uplifted particles demonstrated on the surface of colorectal cells after treatment with new compound 7 for 48 h. Also, complete disruption in cell surface was very clear. In addition, differences in cell three dimensions, increasing average cell roughness and cell diameter due to the effect of compound 7 were illustrated. We suggested in this critical study that sever changes have occurred in the structure of membrane particles accompanied the treatment with compound 7. Additionally, this treatment may lead to apoptosis occurred by remodeling of the membrane skeleton of HCT116 cells. These all morphological changes mentioned above indicated that cell surfaces or membrane compositions (skeleton) were strongly destructed after treatment with our novel compound 7.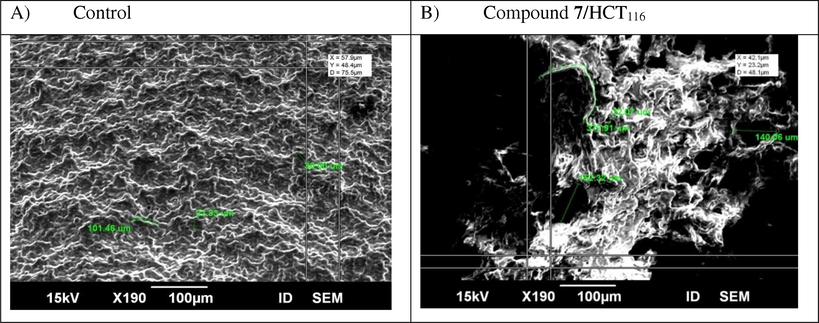
Scanning electron microscope (SEM) images were taken in three dimension and showing the potent apoptotic effect of novel compound 7 at its IC50 value (42.9 µg/ml) on cell surface disruption. A) Untreated colon carcinoma. B) Colon cells treated with compound 7 for 48 h treatment.
3.2.7 TEM microscopic examinations
The specific morphological features of cancer cell death were of the most spreader techniques for quantification and identification of apoptosis, and thus morphologic description using electron microscopy still one of the best tool to identify apoptosis. Transmission electron microscopy (TEM) was used for investigation of apoptosis and necrosis in HCT116 cultured cells, where researchers could study all stages of apoptosis. Data obtained illustrated that control HCT116 cells showed normal structure with a high nucleus-to cytoplasm ratio and a simple cytoplasmic organization with a paucity of organelles. Nucleus, nucleolus and the cytoplasm seemed without abnormal alterations. Mitochondria look like few in number and with its usual elongated form as exhibited in Fig. 8a and c. While Fig. 8b and d showed different apoptotic features as consequence of compound 7 treatment, like cell shrinkage, increase of cytoplasmic organelles, changes of mitochondrial morphology (showed mitochondria with condensed cristae, and mitochondrial membrane rupture as a typical morphological feature of apoptosis), nucleus with reduced volume, chromatin condensation and margination, membrane blebbing and appearance of apoptotic bodies. In addition, (Fig. 8d) clearly showed mitochondrial membrane permialization which promoted the activation of caspases. This permeabilization allowed the release of apoptotic proteins, as cytochrome c and some second mitochondrial activator (DIABLO) from the mitochondrial intermembrane to assembles with apoptotic protease-activating factor-1 (Apaf-1) to induce certain caspase. This caspase, activated the effector caspases which carry out apoptosis. All of these features indicated that compound 7 exerted its antitumor effect against HCT116 cells through the induction of the intrinsic apoptotic pathway.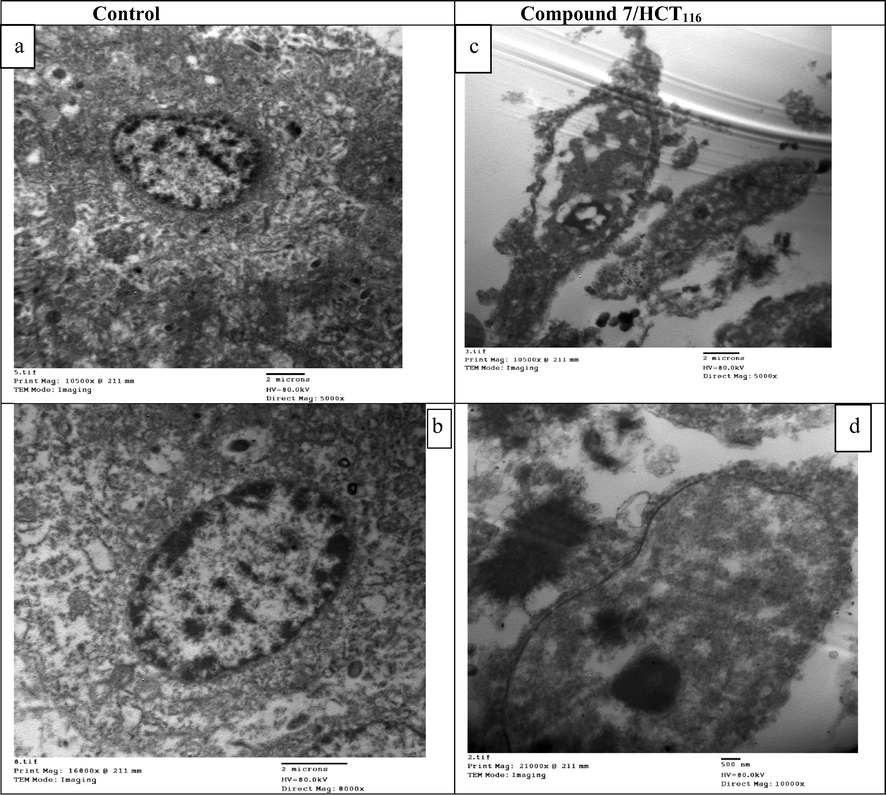
Transmission Electron Microscope (TEM) images of control and compound 7 treated HCT116 cells for 48 hrs of drug incubation. (a and b) Control HCT116 cells showed normal structure patterns with different magnification powers (5000× and 8000×) respectively. (c and d) Compound 7 treated HCT116 cells at different magnification powers (5000× and 1000×) respectively.
3.2.8 Flow cytometer analysis
Signaling transduction Proliferation pathways played a critical role in cancer progression and metastasis, as observed by changing induction and activity of proteins that correlated to the cell cycle progression (Block et al., 2015). Genomic disorder played an important role in cancer progression and initiation. In healthy cells, the genome validity was protected at each step of the cell cycle by investigation at specific points known as checkpoints. In cancerous cells, more than one of these checkpoints could be disrupted. The cellular response to damage may involve activation of a cell cycle checkpoint or when the damage is severe, initiation of apoptosis. Genomic heterogeneity produced might offer uncontrolled tissue growth advantages under certain pressures, such as hypoxia, therapy and immune related provocations. By understanding the mechanism of action of these checkpoints, and how they were overrun in tumor cells, may provide chances for the development of wide spectrum treatment projections (Block et al., 2015). A normal or transformed cell should pass within multiple checkpoints during the division process. Complexes functional proteins were responsible for the operation of these checkpoints that either prevent the cell to proceed or enable it to pass through the checkpoint. Due to DNA disruption, pathways as (ATM and/or ATR) stimulated the activation of a checkpoint that responsible for the arrest or delay that occurred during cell cycle progression. These pathways were outlined by cascade events that were responsible for protein phosphorylation that changed the activity, and the stability of the modified proteins. Here in, inhibition mechanism of compound 7 was further investigated with cell cycle assay and apoptotic study. The novel cyanoacrylamide derivative 7 triggered cell cycle arrest at G2 phase and thus inhibited progression of colon carcinoma cell cycle causing cell death as illustrated in (Table 4 and Fig. 9). There were low decreases in cell population at G1 and S phases of treated sample relative to their control as a result of cell cycle arrest at G2/M phase. The cell cycle arrest at G2/M phase may be triggered by depression in some transcriptional factor responsible for mitotic division, including different cyclin forms and CDC (Medema and Macůrek, 2012), or by downregulation the expression of some proteins responsible for cell cycle progression in G2/M phase. Similar to our study, another cyanoacrylamide derivatives induced cell cycle arrest and apoptosis in breast carcinoma (Mohamed et al., 2017a). The entry of cells into mitosis may be controlled by our target compound through depression the activity of Cdc2 protein. The resulted destabilized protein prevented it from doing its normal function of removing inhibitory phosphorylations from Cdk2. The Cdk2/Cyclin A and Cdk2/Cyclin E complexes still inactive and thus inhibited complete synthesis of DNA and blocked the entry into mitosis. In this assay, it was also observed that compound 7 induced apoptotic cell death of colon carcinoma significantly to reach 12.54% relative to its control (1.84%) as depicted in (Table 5 and Fig. 10). Also, this target 7 induced significant percent of necrosis in HCT116 cells (2.11%) compared to that occurred in control sample (0.42%).
Flow cytometer analysis
Samples
G0/G1 %
S %
G2/M %
Apoptosis %
Control HCT116
58.14
37.04
4.82
1.84
Compound 7/HCT116
43.19
34.41
22.4
12.54
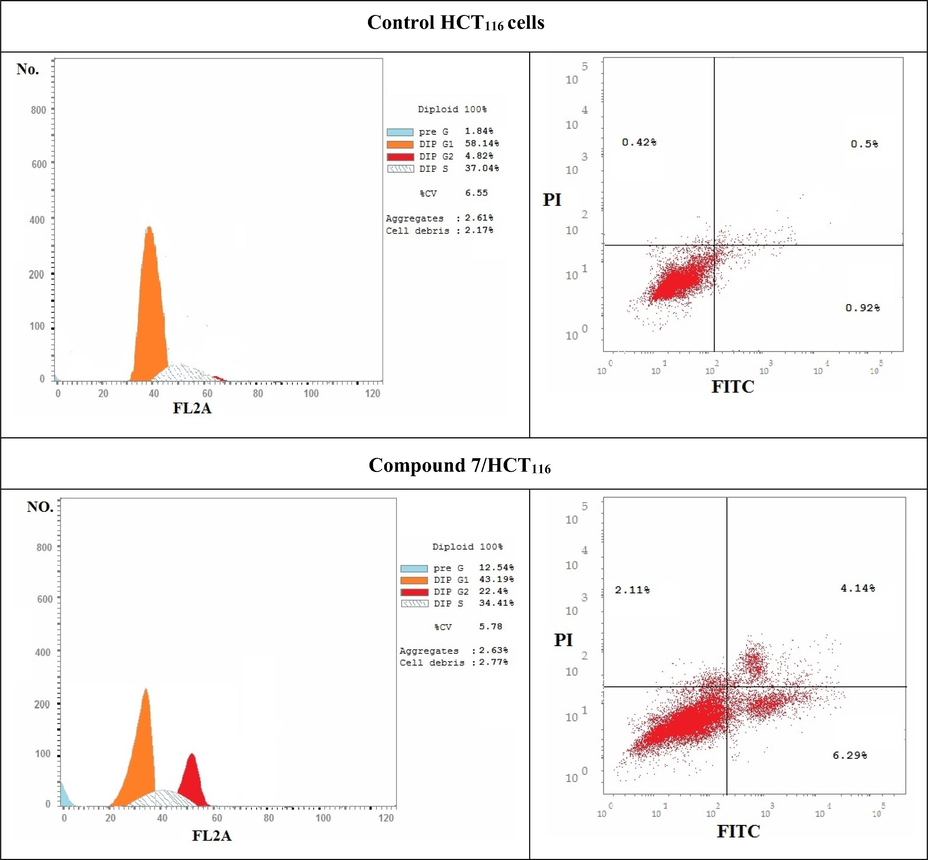
Represent DNA histograms of cell cycle analysis of HCT116 cells after 48 h of treatment with compound 7. The percentage and distribution of cells in each phase of the cell cycles were illustrated.
Samples
Apoptosis %
Necrosis %
%Total
% Early
% Late
Control HCT116
1.84
0.92
0.5
0.42
Compound 7/HCT116
12.54
6.29
4.14
2.11
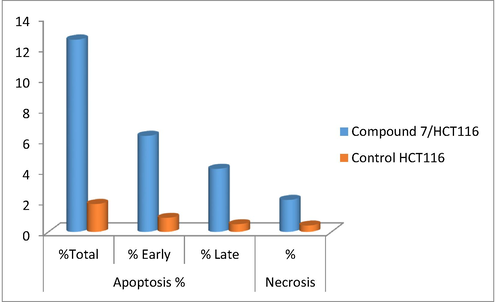
Schematic diagram showed different stages of apoptosis for HCT116 cells treated with compound 7 for 48 h incubation in.
4 Conclusion
In conclusion, it is evident that this study has been suggested a new approaches of synthesis a novel cyanoacylamide incorporating sulphamethoxazol derivative targeting cancer mechanisms of colorectal carcinoma cell line within a wide spectrum approach. This includes combinations of high activity and low toxicity of chemotherapeutic agents, capable of interfering the genesis and metastasis of colorectal carcinoma. New derivatives of cyanoacrylamides were synthesized and screened against different carcinoma cell lines including (A549, HCT116, and MDA) cancer cell lines using MTT method. All tested compounds offered variations in activity, ranging from strong to moderate activities; toward the tested cell lines relative to their lead compound and positive control. Compound 7 among all tested derivatives showed less toxic effect and considered the best cytotoxic compound against all carcinoma, with IC50 values (61.32 µg/ml, 42.9 µg/ml, and 47.5 µg/ml, respectively). Mechanisms of action that compound 7 exerted to inhibit colorectal carcinoma detected using different theoretical and molecular techniques. Giving insight on the binding mode of action of this target ligand, different protein markers (PDB codes: 2wgj, 1dls, 2c6o, 3eyl, 4kmp, 2w3l, 5lab, and 4wt2)) that included in the progression of many carcinomas were tested on it. Compound 7 showed very strong to moderate binding affinity toward all tested domains. It is strongly fit into the active sites of protein markers through hydrogen binding and other types of interactions. Real time PCR analysis showed that upon the treatment of HCT116 cells by compound 7, Caspase-3, BAX, P53 genes were strongly up regulated relative to their control. While BCL2, MMP1, and CDK2 were effectively down regulated suggesting the activation of apoptotic pathway including effective damage of genomic DNA. Flow cytometry assay revealed that compound 7 stimulated cell cycle arrest at G2/M phase, and increased the percentage of cell death more effectively to reach (12.54%). Eliza assay for (caspase-3, cytochrome-c, and VEGF) demonstrated high expression level of caspase-3, and cytochrome-c. While compound 7 decreased the expression level of VEGFR efficiently in the tested line. Finally, morphological investigations were included using (SEM) and (TEM) to observe many changes in the structure and morphology of HCT116 cells which all regulated cell death. Finally, authors suggest that further docking studies confirmed the novel compound exerts its cytotoxic activity by targeting the anti-apoptotic protein markers as (BCL2, MMP, and CDK) which help cancer cells to resist death and continue in progression and metastasis to other organs in the body. Compound 7 performed its action by targeting the active sites of the above protein markers and block them from doing their anti-apoptotic role. In addition, the presence of cyanoacrylamide incorporating both sulphamethoxazole and 3-(thiophen-2-yl)-1H-pyrazole group into the lead compound is responsible for this action. Also, the authors proved that their theoretical studies were compatible to a large extent with their molecular data illustrated in the experimental part. The new inhibitor proved high sensitivity and selectivity toward colorectal carcinoma as cytotoxic agent.
Acknowledgments
Ismail A. Abdelhamid thank the Alexander von Humboldt Foundation, Germany for a postdoctoral fellowship and short-term-visit grants. We thank Prof. Dr. Holger Butenschön, Institut für Organische Chemie, Leibniz Universität Hannover, for fruitful scientific discussion.
Declaration of Competing Interest
All authors declare that they have no conflict of interests.
References
- Carbonic anhydrase inhibitors: benzenesulfonamides incorporating cyanoacrylamide moieties are low nanomolar/subnanomolar inhibitors of the tumor-associated isoforms IX and XII. Bioorg. Med. Chem.. 2013;21:1396-1403.
- [CrossRef] [Google Scholar]
- A novel adamantane thiadiazole derivative induces mitochondria-mediated apoptosis in lung carcinoma cell line. Bioorg. Med. Chem.. 2017;25:241-253.
- [CrossRef] [Google Scholar]
- Antidiabetic activity of aqueous extract of sigesbeckia orientalis (St. paul’s wort) in alloxan-induced diabetes model. Brazilian. J. Pharm. Sci.. 2019;55
- [CrossRef] [Google Scholar]
- A concept of mould spoilage prevention and acrylamide reduction in wheat bread: Application of lactobacilli in combination with a cranberry coating. Food Control. 2018;91:284-293.
- [CrossRef] [Google Scholar]
- Designing a broad-spectrum integrative approach for cancer prevention and treatment. Semin. Cancer Biol.. 2015;35:S276-S304.
- [CrossRef] [Google Scholar]
- Internucleosomal DNA cleavage precedes diphtheria toxin-induced cytolysis. Evidence that cell lysis is not a simple consequence of translation inhibition. J. Biol. Chem.. 1989;264:15261-15267.
- [Google Scholar]
- Designing Smac-mimetics as antagonists of XIAP, cIAP1, and cIAP2. Biochem. Biophys. Res. Commun.. 2009;378:162-167.
- [CrossRef] [Google Scholar]
- Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK) J. Med. Chem.. 2011;54:6342-6363.
- [CrossRef] [Google Scholar]
- Sulfonamide inhibition profile of the β-carbonic anhydrase from Malassezia restricta, an opportunistic pathogen triggering scalp conditions. Metabolites. 2020;10:39.
- [CrossRef] [Google Scholar]
- Swelling kinetics of poly(N-Isopropylacrylamide)-based hydrogel and its application as anti-diabetic drugs delivery system. J. Med. Chem. Sci.. 2019;2:162-171.
- [Google Scholar]
- N-hetaryl-2-cyanoacetamides in the synthesis of substituted (E)-N-hetaryl-2-cyanoacrylamides, (E)-N-alkyl-N-hetaryl-2-cyanoacrylamides, and 6-amino-2-oxo-4-phenyl-1-(pyridin-2-yl)-1,2-dihydropyridine-3,5-dicarbonitriles. Russ. J. Org. Chem.. 2007;43:83-89.
- [CrossRef] [Google Scholar]
- Preparation of some novel 3,5-diaminopyrazole, pyrazolo[1,5-a][1,3,5]triazine and pyrazolo[1,5-a]-pyrimidine derivatives containing sulfonamido moieties as antimicrobial agents. Acta Chim. Slov.. 2002;49:159-171.
- [Google Scholar]
- Studies on aminopyrazoles: antibacterial activity of some novel pyrazolo[1,5-a]pyrimidines containing sulfonamido moieties. Farm. 2000;55:596-602.
- [CrossRef] [Google Scholar]
- Novel synthesis and antifungal activity of pyrrole and pyrrolo[2,3-d]pyrimidine derivatives containing sulfonamido moieties. Farm. 2002;57:613-617.
- [CrossRef] [Google Scholar]
- Synergistic interactions of sulfamethoxazole and azole antifungal drugs against emerging multidrug-resistant Candida auris. Int. J. Antimicrob. Agents. 2018;52:754-761.
- [CrossRef] [Google Scholar]
- Utility of activated nitriles in the synthesis of some new heterocyclic compounds. Am. J. Org. Chem.. 2012;22:32-40.
- [CrossRef] [Google Scholar]
- Design, synthesis and anticancer evaluation of novel tetrahydroquinoline derivatives containing sulfonamide moiety. Eur. J. Med. Chem.. 2009;44:4211-4217.
- [CrossRef] [Google Scholar]
- Cytotoxic and antimicrobial evaluations of novel apoptotic and anti-angiogenic spiro cyclic 2-oxindole derivatives of 2-amino-tetrahydroquinolin-5-one. Arch. Pharm. (Weinheim). 2015;348:113-124.
- [CrossRef] [Google Scholar]
- Rapid microwave-assisted solution phase synthesis of substituted 2-pyridone libraries. Tetrahedron. 2004;60:8633-8644.
- [CrossRef] [Google Scholar]
- Combination of sulfamethoxazole and selenium in anticancer therapy: a novel approach. Mol. Cell. Biochem.. 2013;384:279-285.
- [CrossRef] [Google Scholar]
- Anti-fungal Activity of Sulfamethoxazole toward Aspergillus Species. Biol. Pharm. Bull.. 2005;28:773-778.
- [CrossRef] [Google Scholar]
- Electron microscopic analysis of different cell types in human pancreatic cancer spheres. Oncol. Lett.. 2017;15:2485-2490.
- [CrossRef] [Google Scholar]
- Synthesis of a novel p-hydroxycinnamic amide with anticancer capability and its interaction with human serum albumin. Exp. Ther. Med.. 2019;17:1321-1329.
- [CrossRef] [Google Scholar]
- Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res.. 1993;53:3976-3985.
- [Google Scholar]
- Methotrexate-resistant variants of human dihydrofolate reductase with substitutions of leucine 22. Kinetics, crystallography, and potential as selectable markers. J. Biol. Chem.. 1995;270:5057-5064.
- [CrossRef] [Google Scholar]
- Microwave-assisted preparation of biodegradable, hemocompatible, and antimicrobial neem gum–grafted poly (acrylamide) hydrogel using (3)2 factorial design. Emergent Mater.. 2019;2:95-112.
- [CrossRef] [Google Scholar]
- Relations between structure and biological activity of sulfonamides. Annu. Rev. Pharmacol. Toxicol.. 1976;16:309-327.
- [Google Scholar]
- Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci.. 2000;97:10389-10394.
- [CrossRef] [Google Scholar]
- ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev.. 2001;15:1067-1077.
- [CrossRef] [Google Scholar]
- A randomized trial of short-course ciprofloxacin, ofloxacin, or trimethoprim/sulfamethoxazole for the treatment of acute urinary tract infection in women. Am. J. Med.. 1999;106:292-299.
- [CrossRef] [Google Scholar]
- Anticancer Activities of New N-hetaryl-2-cyanoacetamide Derivatives Incorporating 4,5,6,7-Tetrahydrobenzo[b]thiophene Moiety. Anticancer. Agents Med. Chem.. 2017;17:1084-1092.
- [CrossRef] [Google Scholar]
- Cytotoxicity, molecular modeling, cell cycle arrest, and apoptotic induction induced by novel tetrahydro-[1,2,4]triazolo[3,4-a]isoquinoline chalcones. Eur. J. Med. Chem.. 2018;143:532-541.
- [CrossRef] [Google Scholar]
- Chalcones incorporated pyrazole ring inhibit proliferation, cell cycle progression, angiogenesis and induce apoptosis of MCF7 cell line. Anticancer. Agents Med. Chem.. 2014;14:1282-1292.
- [Google Scholar]
- Synthesis and biological evaluation of a novel series of chalcones incorporated pyrazole moiety as anticancer and antimicrobial agents. Appl. Biochem. Biotechnol.. 2012;168:1153-1162.
- [CrossRef] [Google Scholar]
- Apoptotic induction mediated p53 mechanism and Caspase-3 activity by novel promising cyanoacrylamide derivatives in breast carcinoma. Bioorg. Chem.. 2017;73:43-52.
- [CrossRef] [Google Scholar]
- Synthesis and characterization of new heterocyclic (triazoline) anticancer compounds. Indian J. Public Heal. Res. Dev.. 2019;10:1121-1124.
- [CrossRef] [Google Scholar]
- New acrylamide-sulfisoxazole conjugates as dihydropteroate synthase inhibitors. Bioorg. Med. Chem.. 2020;28:115444.
- [CrossRef] [Google Scholar]
- Empiric use of trimethoprim-sulfamethoxazole (TMP-SMX) in the treatment of women with uncomplicated urinary tract infections, in a geographical area with a high prevalence of TMP-SMX–resistant uropathogens. Clin. Infect. Dis.. 2002;34:1165-1169.
- [CrossRef] [Google Scholar]
- Design, synthesis, and antifungal activity of novel cinnamon-pyrazole carboxamide derivatives. Drug Dev. Res.. 2018;79:307-312.
- [CrossRef] [Google Scholar]
- Triazolo[1,5-a]pyrimidines as novel CDK2 inhibitors: Protein structure-guided design and SAR. Bioorg. Med. Chem. Lett.. 2006;16:1353-1357.
- [CrossRef] [Google Scholar]
- Roifman, C., Aviv, G., Alexander, L., 2000. Preparation of N-benzyl-3-aryl-2-cyanoacrylamides activity for treatment of neoplastic disorders. PCT. Int. Appl. WO Pat. PCT. Int. Appl. WO Patent 2000, 0055, 128.
- The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev.. 2000;14:289-300.
- [CrossRef] [Google Scholar]
- Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem.. 2002;277:46265-46272.
- [CrossRef] [Google Scholar]
- Synthesis and Spectral Properties of 1-(6-Methoxy-2-benzothiazolyl)-2-pyridones. Collect. Czech. Chem. Commun.. 1996;61:921-929.
- [Google Scholar]
- In vitro assessment of antifungal drugs and sulfamethoxazole-trimethoprim against clinical isolates of conidiobolus lamprauges. Antimicrob. Agents Chemother.. 2018;62:e01685-e1717.
- [CrossRef] [Google Scholar]
- Effects of the antimicrobial sulfamethoxazole on groundwater bacterial enrichment. Environ. Sci. Technol.. 2011;45:3096-3101.
- [CrossRef] [Google Scholar]
- Antimicrobial activities of trimethoprim/sulfamethoxazole, 5-iodo-2′-deoxyuridine and rifampicin against Staphylococcus aureus. Int. J. Antimicrob. Agents. 2010;36:562-565.
- [CrossRef] [Google Scholar]
- A Canadian national surveillance study of urinary tract isolates from outpatients: comparison of the activities of trimethoprim-sulfamethoxazole, ampicillin, mecillinam, nitrofurantoin, and ciprofloxacin. The Canadian Urinary Isolate Study Group. Antimicrob. Agents Chemother.. 2000;44:1089-1092.
- [CrossRef] [Google Scholar]
- Discovery of trans-3-(pyridin-3-yl)acrylamide-derived sulfamides as potent nicotinamide phosphoribosyltransferase (NAMPT) inhibitors for the potential treatment of cancer. Bioorg. Med. Chem. Lett.. 2019;29:1502-1506.
- [CrossRef] [Google Scholar]
- A simplified diphenylamine colorimetric method for growth quantification. Appl. Microbiol. Biotechnol.. 2013;97:5069-5077.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2020.04.032.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







