Translate this page into:
A novel ion mobility separation-enabled and precursor ions list-included high-definition data-dependent acquisition (HDDDA) approach: Method development and its application to the comprehensive multicomponent characterization of Fangji Huangqi Decoction
⁎Corresponding authors at: State Key Laboratory of Component-based Chinese Medicine, Tianjin University of Traditional Chinese Medicine, 10 Poyanghu Road, Jinghai, Tianjin 301617, China. gaoxiumei@tjutcm.edu.cn (Xiumei Gao), wzyang0504@tjutcm.edu.cn (Wenzhi Yang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
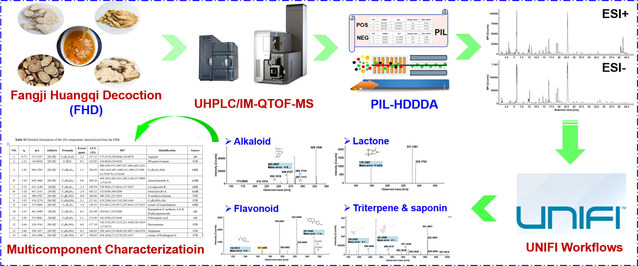
To elucidate the chemical composition of a traditional Chinese medicine (TCM) formula necessitates the development of more potent analytical strategies, because of the complexity as a result of the superposition of multiple drugs. Fangji Huangqi Decoction (FHD) is a four-component TCM formula composed by complicated chemical constituents. By using the VionTM ion mobility quadrupole time-of-flight (IM-QTOF) mass spectrometer, we present a novel IM separation-enabled and precursor ions list (PIL)-included high-definition data-dependent acquisition (HDDDA) approach, and apply it to the multicomponent characterization of FHD by coupling to ultra-high performance liquid chromatography. Chromatographic separation was conducted on a CORTECS® UPLC® T3 column, while HDDDA was employed for MS2 data acquisition in both the negative and positive electrospray-ionization (ESI) modes. The PILs of FHD in two ESI modes were created based on the phytochemical knowledge of four component drugs and mass defect filtering, which ultimately could obtain 316 and 258 targeted masses, respectively, from the full-scan MS1 data. Interestingly, the additional inclusion of PILs in HDDDA could improve the coverage on the target components by 12% (ESI–) and 48% (ESI + ). Structural elucidation was performed by in-house database-driven automatic peak annotation using UNIFITM. We could identify or tentatively characterize 203 components from FHD, involving 25 alkaloids, 86 flavonoids, 48 triterpenoids (saponins), 16 lactones, and 28 others). It is the first report regarding the method development of HDDDA that targets the global TCM components characterization, and the findings obtained would benefit the quality control and the secondary development of FHD.
Keywords
Fangji Huangqi Decoction
Multicomponent characterization
Ion mobility/quadrupole time-of-flight mass spectrometry
Precursor ions list
High-definition data-dependent acquisition
1 Introduction
The principle of traditional Chinese medicine (TCM) is “differentiation and treatment”, and a TCM formula is formulated based on the compatibility theory (“Monarch, Minister, Assistant, and Guide”) according to the different syndromes. TCM formulae refer to the prescriptions consisting of two or more drugs, which are the main form of TCM for the clinical use (Zhou et al., 2017a). The main characteristics of TCM formulae are the integrity and complexity. On one hand, the chemical composition of TCM formulae is extremely complex (Guo et al., 2015), which may result from two factors: 1) complicated chemical composition of the single component drug (e.g. the co-existing of the primary and secondary metabolites of plants, and > 600 kinds of saponins could be characterized from the leaf of Panax ginseng (Qiu et al., 2015; Yan et al., 2017), and 2) the superposition of multicomponents from all the component drugs, and even their interactions to yield new substances (Xu et al., 2017). On the other hand, the mechanism of action for TCM formulae trends to be more complex, such as the interaction and synergistic action among different ingredients contained (Liu et al., 2018). A consensus has been reached that, the dimness in the chemical composition of TCM formulae acts as a key obstacle involved in the process of TCM modernization and globalization, which directly restricts the investigations with respect to the quality control, pharmacology, and mechanism of action, etc.
Rapid development of the analytical technologies, especially liquid chromatography-mass spectrometry (LC-MS), has greatly driven and facilitated the comprehensive multicomponent characterization of natural products (Stavrianidi, 2020). On one hand, the multi-dimensional liquid chromatography (MDLC), operating in either the on-line or off-line mode, greatly improves the peak capacity and enhances the selectivity of separation, particularly when orthogonal chromatography is used (Fu et al., 2019; Sagirli et al., 2019; Zhou et al., 2020). In contrast, the on-line mode enables high automation and is easy to standardize, while the off-line pattern is more flexible and easily popularized among laboratories. On the other hand, versatile MS scan methods and fragmentation mechanisms are currently available on the advanced mass spectrometers. For instance, data-independent acquisition (DIA) and data-dependent acquisition (DDA) strategies in an untargeted mode are widely applied in the profiling and characterization of plant metabolites and TCM components. Comparatively, DIA is an unbiased scan approach receiving a complete coverage of all the precursors, by allowing the fragmentation of the ions within splitted and continuous mass windows (namely Sequential Window Acquisition for All Theoretical Spectra-SWATH for AB SCIEX) or the entire scan range (e.g. MSE for Waters, All Ions Fragmentation-AIF for Thermo Fisher Scientific, and All Ions MS/MS for Agilent). However, spectral deconvolution, to match the precursor-product ions, is inevitable prior to the data interpretation for DIA strategies (Huh et al., 2019; Kinyua et al., 2015; Navarro-Reig et al., 2017; Xia et al., 2019), which can be performed either by the commercial software (such as UNIFITM from Waters) or in-house developed algorithms (e.g. MS-DIAL (Tsugawa et al., 2015), MetDIA (Li et al., 2016), and DecoMetDIA (Yin et al., 2019), etc.). On occasion, the precursor-product ion matching results may contain false positives because of the presence of adducts or in-source decay fragments. In general, DDA remains as the most commonly used strategy due to its easy accessibility, by which no prior knowledge is required ahead of the analysis and the data processing is easy to handle for the structural elucidation. However, while facing the remarkable interferences, the intensity-ranking criteria to automatically select the top N most intense precursor ions for triggering MS2 by DDA can result in very low coverage on the components of interest (Wang et al., 2019). As a solution to this issue, a precursor ions list (PIL) consisting of the target masses or an exclusion list (EL) composed by the unwanted masses can be set in DDA, by which the sensitivity in charactering the target components can be significantly improved (Li et al., 2020b; Pan et al., 2020). Another marked advancement in TCM component analysis is known as the introduction of ion mobility mass spectrometry (IM-MS), and the commercial instruments involve the SYNAPT series (HDMS, G2 HDMS, G2-Si HDMS, and XS), Vion IM-QTOF, SELECT SERIES Cyclic IMS from Waters, and 6560 IM-QTOF from Agilent. Hybrid IM-QTOF-MS coupled to LC provides the four-dimensional information related to each analyte (involving tR, drift time, MS1, and MS2) (Li et al., 2020a; Wang et al., 2020). More importantly, IM-derived collision cross section (CCS) can be utilized for database elaboration in support of the metabolites identification and even the isomers differentiation (Tu et al., 2019; Zhou et al., 2016; Zhou et al., 2017b). Standardized workflows enabling the automatic peak annotation of the high-definition MSE (HDMSE) and DDA data have been established by using the Vion IM-QTOF and UNIFI, which can demonstrate an efficient and intelligentized strategy facilitating the dimension-enhanced, more reliable metabolites characterization (Koley et al., 2020; Zhang et al., 2019; Zuo et al., 2019).
Fangji Huangqi Decoction (FHD) is a famous TCM formula composed by Stephaniae Tetrandrae Radix (STR; Fang-Ji), Astragali Radix (AR; Huang-Qi), Atractylodis Macrocephalae Rhizoma (AMR; Bai-Zhu), and Glycyrrhizae Radix et Rhizoma (GRR, Gan-Cao). It has the benefits of “tonifying qi to expel wind and invigorating spleen for diuresis”, which can be prescribed to treat the chronic glomerulonephritis, cardiogenic edema, and rheumatoid arthritis embolism (Wang et al., 2016a). The advanced LC-MS approaches have been applied to qualitative and quantitative analyses of the multicomponents of FHD together with the prototype compounds and metabolites in rat plasma (Wang et al., 2015a; Wang et al., 2015b; Wang et al., 2016b). A UHPLC/QTOF-MS approach, by feat of dynamic background subtraction and enhanced peak list data processing, was able to characterize 55 components from FHD, 8 prototypes, and 33 metabolites. Unfortuantely, the in-depth chemical basis elucidation of FHD has not been reported, hitherto.
In the current work, we introduce a novel analytical strategy, by integrating IM-enabled and PIL-included high-definition DDA (namely HDDDA) coupled to the ultra-high performance reversed-phase liquid chromatography (RP-UHPLC) and the computational peak annotation, as a solution to facilitate the in-depth multicomponent characterization of FHD. Notably, the HDDDA represents a novel IM separation based DDA strategy able to generate the MS spectra with less interference. To boost the coverage and simultaneously streamline the entire workflows, the following efforts have been made: i) optimizing both the chromatography condition and the key parameters of the VionTM IM-QTOF mass spectrometer; ii) the utilization of HDDDA in two ESI modes (ESI–/ESI + ) to generate high-definition and more reliable MS/MS spectra; iii) the inclusion of PILs in HDDDA generated by the phytochemical knowledge and mass defect filtering; iv) establishment of the standardized workflows for database-driven computational peak annotation by using UNIFI; and v) the use of more reference compounds (62 compounds in total) and four single drugs (STR, AR, AMR, and GRR) to elevate the reliability of structural elucidation. Despite HDDDA having been recently reported for the purpose of metabolites characterization (Radchenko et al., 2020), the method development workflows by applying HDDDA to comprehensively characterize the multicomponents from a complex herbal or biofluid sample in an untargeted mode are reported for the first time. An in-house FHD library could guide the generation of PILs by mass defect filtering and the “Identified Compounds” list by UNIFI. As a consequence, the sufficient resolution and sensitive characterization of versatile ingredients from FHD were achieved with a number of components newly exposed, characterized, and the provision of CCS values, which can set an example for the in-depth multicomponent characterization of a complex sample like the TCM formula.
2 Materials and methods
2.1 Reagents and chemicals
In total, 62 compounds (the chemical structures are given in Fig. 1 and the detainled information offered in Table S1, as Supplementary Materials) involving 16 flavonoids (numberred as 1–16), 15 alkaloids (17–31), 7 saponins (32–38), and 24 others (39–62), purchased from Shanghai Standard Biotech. Co., Ltd. (Shanghai, China) or Chengdu Desite Biotechnology Co., Ltd. (Chengdu, China), were used as the reference compounds in this work. Acetonitrile, methanol (Fisher, Fair lawn, NJ, USA), formic acid (FA), and ammonium acetate (Sigma-Aldrich, St. Louis, MO, USA), were all the LC-MS grade. Ultra-pure water was prepared using a Milli-Q Integral 5 water purification system (Millipore, Bedford, MA, USA). Information of the decoction pieces that had been used to prepare the FHD sample was as follows. STR: Jinggongqiao Town, Fuliang County, Jingde Town, Jiangxi Province, AR: Ningxia Hui Autonomous Region; AMR: Pan'an, Zhejiang Province; GRR: Yanchi County, Ningxia Hui Autonomous Region. All samples were deposited in the authors’ laboratory, Tianjin University of Traditional Chinese Medicine (Tianjin, China).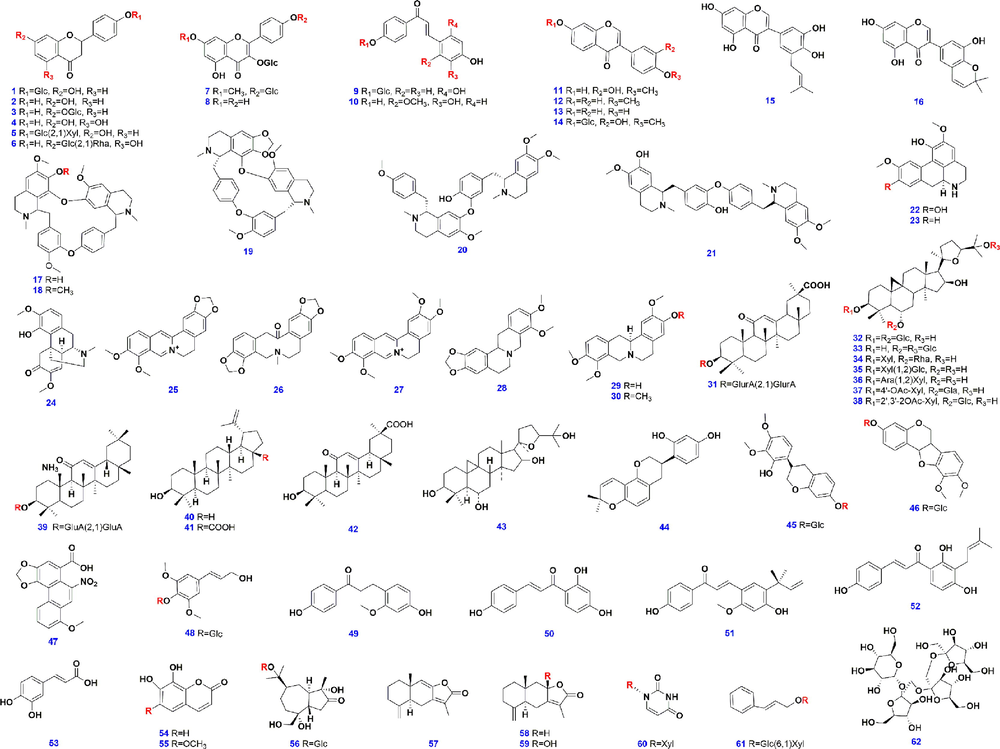
Chemical structures of the 62 reference compounds.
2.2 Sample preparation
The FHD sample was prepared by the reflux extraction. In detail, 15 g of AR, 12 g of STR, 9 g of AMR (braised), and 6 g of GRR, were extracted for 2.5 h under reflux by 8-fold the amount of water (336 mL). After filtering, the residue was extracted for the second time using the same volume of water for 1.5 h, and then filtered. The obtained extract liquids were pooled, and further diluted with pure water reaching the concentration of 20 mg/mL. After centrifugation at 11,481 g (equal to 14,000 revolutions per minute) for 10 min, the supernatant was treated as the test solution of FHD. To prepare the samples of STR, AR, AMR, and GRR, each drug (10.0 g) was accurately weighed and extracted using the same method, described for extracting FHD, with the final concentration of 20 mg/mL. The same centrifugation approach was employed and the test solutions for STR, AR, AMR, and GRR, were obtained by taking the supernatant. To the 62 reference compounds, an aliquot of 1.0 mg for each was accurately weighed and dissolved in methanol yielding a solution at the concentration of 200 μg/mL as the stock solutions. The equal volume of each of the reference compound test solution was taken to prepare four mixed standard samples. They were centrifuged at 11,481 g for 10 min, and the supernatants obtained were used as the test solutions for the reference compounds.
2.3 UHPLC/IM-QTOF-MS using HDDDA
The multicomponent profiling and characterization of FHD was performed by an ACQUITY UPLC I-Class/VionTM IM-QTOF system (Waters Corporation, Milford, MA, USA). A CORTECS® UPLC® T3 column (2.1 × 100 mm, 1.6 µm) maintained at 30 °C was used for the chromatographic separation. A binary mobile phase, composed by 0.1% FA in H2O (water phase: A) and 0.1% FA in CH3CN (organic phase: B), was employed at a flow rate of 0.3 mL/min following an optimal gradient program: 0–1 min, 1% (B); 1–2 min, 1–15% (B); 2–7 min, 15–20% (B); 7–14 min, 20–30% (B); 14–24 min, 30–35% (B); 24–30 min, 35–40% (B); 30–36 min, 40%–58% (B); 36–39 min, 58–74% (B); 39–39.5 min, 74–95% (B); and 39.5–41.5 min, 95% (B). A 5-min re-equilibration time was set between the successive injections. The injection volume was set at 3 µL.
High-accuracy MS data were recorded on a Vion IM-QTOF mass spectrometer by HDDDA (High Definition MS → High Selectivity MS/MS) in both the negative and positive ESI modes. A LockSpray ion source was equipped using the following parameters: capillary voltage, –2.5 kV/2.5 kV; cone voltage, 80 V; source offset, 80 V; source temperature, 120 °C; desolvation gas temperature, 500 °C; desolvation gas flow (N2), 800 L/h; and cone gas flow (N2), 50 L/h. Default parameters were defined for the travelling wave IM separation. The QTOF analyzer scanned over a mass range of m/z 100–1000 at a low energy of 6 eV for both two ESI modes at 0.3 s per scan for MS1 profiling, and over the same mass range at 0.2 s per scan for recording the MS2 data. In the positive mode, when TIC (total ion chromatogram) intensity exceeded 1000 detector counts, the MS/MS fragmentation of three most intense precursors was automatically triggered, and mass-dependent ramp collision energy (MDRCE, widely known as a mass-specific collision energy strategy) of constant 15 eV in low mass ramp and 35 eV in high mass ramp was set for the positive-mode HDDDA. The MS/MS acquisition stopped if either TIC exceeded 500 detector counts or time exceeded 0.8 s. In the negative-mode HDDDA, when the TIC intensity exceeded 1000 detector counts, the MS/MS fragmentation of the four most intense precursors was automatically triggered, and MDRCE of constant 15 eV in low mass ramp and 35 eV in high mass ramp was set. The MS/MS acquisition stopped if either TIC exceeded 1000 detector counts or time exceeded 1.0 s. Detained settings for the PILs in the positive and the negative modes are given in Table S2 and Table S3, respectively. A solution of leucine enkephalin (Sigma-Aldrich, St. Louis, MO, USA; 200 ng/mL) was constantly infused at a flow rate of 10 µL/min. Calibration of CCS was conducted according to the manufacture’s guideline using a mixture of calibrants (Paglia et al., 2015).
2.4 Data processing by UNIFI
The uncorrected collision-induced dissociation (CID)-MS2 data obtained by HDDDA (in the continuum format) were processed by the UNIFITM software, which performed data correction, peak picking, and peak annotation by searching against an incorporated in-house FHD library. Key parameters of UNIFI are depicted as follows: DDA Mass Detection: MS ion intensity threshold, 1,500.0 counts; MS/MS ion intensity threshold, 600.0 counts; Maximum number of MS ions to keep per setMass, 5,000; Maximum number of MS/MS ion to keep per setMass, 5,000. Target by mass: target match tolerance, 10.0 ppm; screen on all isotopes in a candidate; generate predicted fragments from structure, fragment match tolerance, 10.0 ppm; If more than one target component is assigned to a candidate then display only the best matching target; Maximum candidates per sample to use during screening and discovery, 50,000; Maximum candidates per sample to keep after screening and discovery, 50,000. Adducts: positive adducts including + H, +Na, +NH4, +K; negative adducts including + HCOO, –H, +CH3COO, +Cl. Lock Mass Settings: combine width, 3 scans; mass window, 0.5 m/z; reference mass, positive: 556.2766; negative: 554.2620; reference charge, +1/–1.
3 Results and discussion
3.1 Optimization of a UHPLC/IM-QTOF-MS approach dedicated to the global chemical profiling of FHD
Because of the combination of four herbal medicines (STR, AR, AMR, and GRR), the chemical composition of FHD becomes more complex. In this context, more potent analytical strategies, which can well resolve and sensitively identify the multiple classes of herbal components contained, are in great need. The Vion IM-QTOF hybrid high-resolution mass spectrometer by couppling to UHPLC (LC, IM, and MS) can enable the three-dimensional separation, rendering a powerful platform appliable to the comprehensive chemical basis elucidation of TCM (Wang et al., 2020; Zhang, et al., 2019; Zhang et al., 2020; Zuo et al., 2019). In the present work, we develop an IM-enabled and PILs-included HDDDA approach, aiming to deeply expose and characterize the multicomponents from FHD. For this purpose, diverse efforts were made to improve the performance of both UHPLC separation and the detection by Vion IM-QTOF-MS. Considering the differentiated ionization properties for the multiple classes of natural components (such as the saponins, flavonoids, alkaloids, and terpenes, etc.) present in FHD (Wang et al., 2015b), both the negative and positive ESI analyses were employed.
Key segments of RP-UHPLC were optimized in the first step. We selected formic acid (FA) as the additive in the water phase, under which both the neutral, acidic, and basic compounds got well separated. The columns prepared with different bonding technology or bonding groups from different vendors may exert significantly different resolution for the constituents under study (Fu et al., 2019; Koley et al., 2020; Zhang et al., 2019; Zuo et al., 2019). We evaluated the effects of ten different RP-UHPLC columns (from three vendors) on the separation of FHD components by comparing the overall resolution and the number of the resolvable peaks eluted using a uniform gradient elution procedure. These candidate columns include CORTECS UPLC T3, CSH C18, BEH Shield RP18, BEH C18, HSS T3, CSH Phenyl-Hexyl, and CSH Fluoro-Pheniyl from Waters, ZORBAX SB C18 and ZORBAX Eclipse Plus C18 from Agilent, and Kinetex XB C18 from Phenomenex. As a result, except the CSH Phenyl-Hexyl and CSH Fluoro-Pheniyl columns bonded with the phenyl groups (extremely bad resolution was observed), all the other eight columns were considerred as suitable which could be utilized to analyze the multicomponents from FHD (Fig. 2). Comparatively, CORTECS UPLC T3 could resolve the largest number of peaks (12301) and showed certain retention of the polar components (tR < 5 min), which indicated very high selectivity against the complex components of FHD. It combines the merits of T3 bonding technology and the core–shell particles, and can endure 100% aqueous mobile phase with high column efficiency. Therefore, we chose the CORTECS UPLC T3 column in this experiment. Next, the effects of column temperature (varying between 25 and 35 °C) were evaluated by using the selected column. Obviously, an increase in temperature could enhance the elution ability, under which the retention of some components was weakened. The column temperature was set at 30 °C in the subsequent experiments. Moreover, slight adjustment on the elution gradient was made, which, as a result, resulted in good separation performance with the multicomponents from FHD well resolved.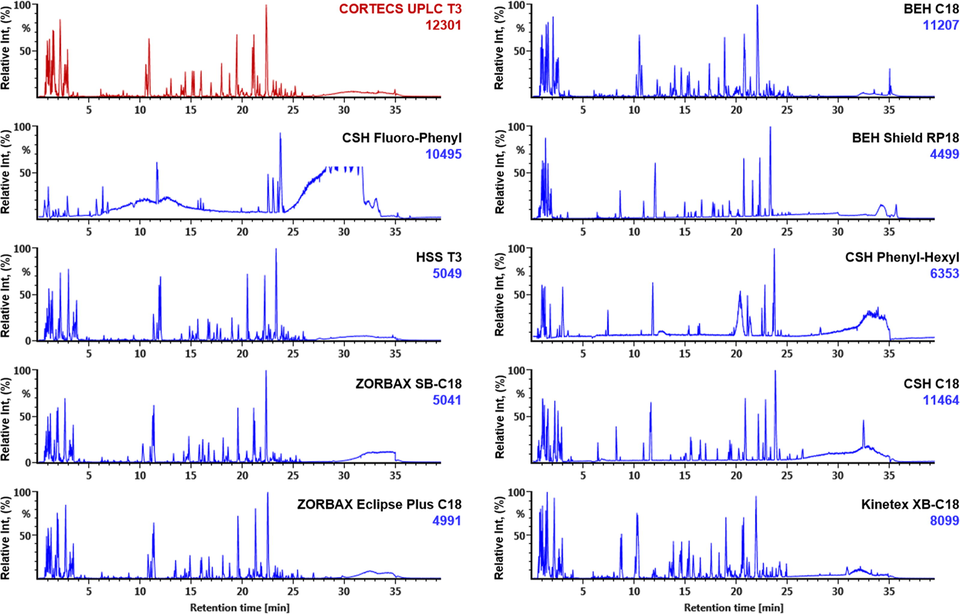
Selection of the stationary phase for the reversed-phase UHPLC separation of the multicomponents from FHD by evaluating ten different sub-2 µm particles packed chromatographic columns purchased from three vendors (Waters, Agilent, and Phenomenex). The base peak intensity chromatograms are obtained in the negative high-definition MS mode. The numbers marked in blue (and red) refer to the peaks resolved deduced by processing the MS1 data using UNIFI.
Key parameters of the Vion IM-QTOF instrument, which could influence the overall ion response (MS1) and the fragmentation degree of the precursor ions (MS2), and data-dependent CID-MS2 acquisition, were optimized in both the positive and negative ESI modes. Two ion source parameters, capillary voltage (varying in the range of 1.0–3.0 kV) and cone voltage (20–100 V), were optimized by measuring the variations of the peak areas of the precursor ions. Considering the chemical diversity of FHD, ten reference compounds, involving calycosin and formononetin (flavonoids), (+)-norisoboldine, tetrandrine, and fangchinoline (alkaloids), and atractylenolide I (lactone), in the positive ESI mode, and liquiritin and neoisoliquiritin (flavonoid glycosides), astragaloside IV and glycyrrhizic acid (saponins) in the negative mode, were taken as the index compounds (Fig. 3). In the positive mode, four of the six index compounds were best ionized at capillary voltage of 2.5 kV, while the other two (tetrandrine and fangchinoline) gave the highest ion response at 3.0 kV (slight increase compared with that at 2.5 kV). We thus selected the capillary voltage of 2.5 kV in the positive mode. In addition, the changes of cone voltage ranging 20–100 V led to the inconsistent variation trends for the index compounds. The ion response for (+)-norisoboldine (a minor component in FHD) showed sharp decreasing when cone voltage was set higher than 40 V. However, the majority of these index compounds could be better ionized when a high level of cone voltage was set (40–80 V). Collectively, we selected the cone voltage of 80 V in the positive mode. In the case of the negative mode, the best ionization was observed at capillary voltage of 2.5 kV for three compounds and at 3.0 kV for one compound (glycyrrhizic acid), respectively. Therefore, the capillary voltage in the negative mode was set at 2.5 kV. Cone voltage in the negative mode also exerted different effects on the peak areas for different index compounds. A high level of cone voltage at 80 V was selected in the negative ion mode, under which the flavonoid glycoside liquiritin and neoisoliquiritin, as well as the saponin astragaloside IV, showed the highest peak area values and the second highest for the other saponin compound, glycyrrhizic acid. It was noted that, despite that the weak (or medium) in-source fragmentation was observed for some flavonoid compounds in the negative mode and the flavonoids and saponins in the positive mode, it could seldom influence their characterization because of the in-house library-driven compurational characterization strategy (including a confirming process to remove the false positive characterizations) we apply. Moreover, to enable the sufficient fragmentation for diverse classes of components from FHD, we selected to apply mass-dependent ramp collision energy (MDRCE), by which mass-specific ramp collision energy can be achieved (Zhang et al., 2019). Using the FHD sample, the CID-MS2 spectra obtained at five different levels of MDRCE in the negative ESI mode were compared, by setting fixed voltages both at the low mass and high mass regions (low mass: 10 eV/high mass: 30 eV, 15/35 eV, 20/40 eV, 25/45 eV, and 30/50 eV). As the compounds detectable in the positive ion mode mainly involved the alkaloids from STR and lactones from AMR, we assessed these different levels of MDRCE settings based on the numbers of components that could be automatically identified via matching the in-house libraries of STR and AMR. As a consequence, the numbers of components automatically identified were 37, 51, 44, 42, and 41, based on the CID-MS2 data obtianed at five levels or ramp collision energy, respectively, which could suggest a good choice of MDRCE at 15 eV/35 eV in the positive mode. In the negative ESI mode, we selected three compounds as the indicators (liquiritin apioside, neoisoliquiritin, and glycyrrhizic acid), to compare the same five levels of MDRCE. As a result, the similar trends as those in the positive mode were witnessed, and thereby we set the same MDRCE (15 eV/35 eV). Moreover, we optimized the MS/MS parameters which guided the automatic switching between the MS1 and MS2 acquisitions, by using the number of “Identified Components” as the indicator (automatic peak annotation by UNIFI generating the list of the primarily identified compounds). The threshold of TIC (detector counts) drove the switching from MS1 to MS2, while either TIC threshold or timeout guided the switching to the full-scan MS1 acquisition. In the positive ion mode, the TIC threshold was set at 100, 500, and 1000 counts, and accordingly, the numbers of those identified components were 371, 375, and 352 (the raw characterization result prior to the confirming process), by calling the databases of four component drugs. In addition, four groups of top N settings (top N/timeout) related to stopping the MS/MS acquisition (scan time of 0.2 s), including top 1/0.3 s, top 2/0.5 s, top 3/0.8 s, and top 4/1.0 s, were compared in this section. And eventually, the numbers of the identified compounds corresponded to 40, 214, 493, and 479, respectively. Therefore, in the positive ESI mode, we chose the settings of TIC threshold of 500 counts to trigger the MS/MS acquisition and top 3/timeout 0.8 s to automatically stop the MS/MS scan. In the negative mode, the TIC threshold at five levels (100, 500, 1000, 1500, and 2000), and the numbers of those automatically identified components were 463, 467, 479, 477, and 452, respectively. The four levels of top N/timeout settings could result in the identification of 210, 233, 416, and 468 components, respectively. Taking together, in the negative ion mode, we finally set TIC threshold at 1000 to trigger the MS/MS acquisition and top 4/timeout 1.0 s to automatedly terminate the MS/MS scan. These optimal parameters were set to establish the HDDDA methods.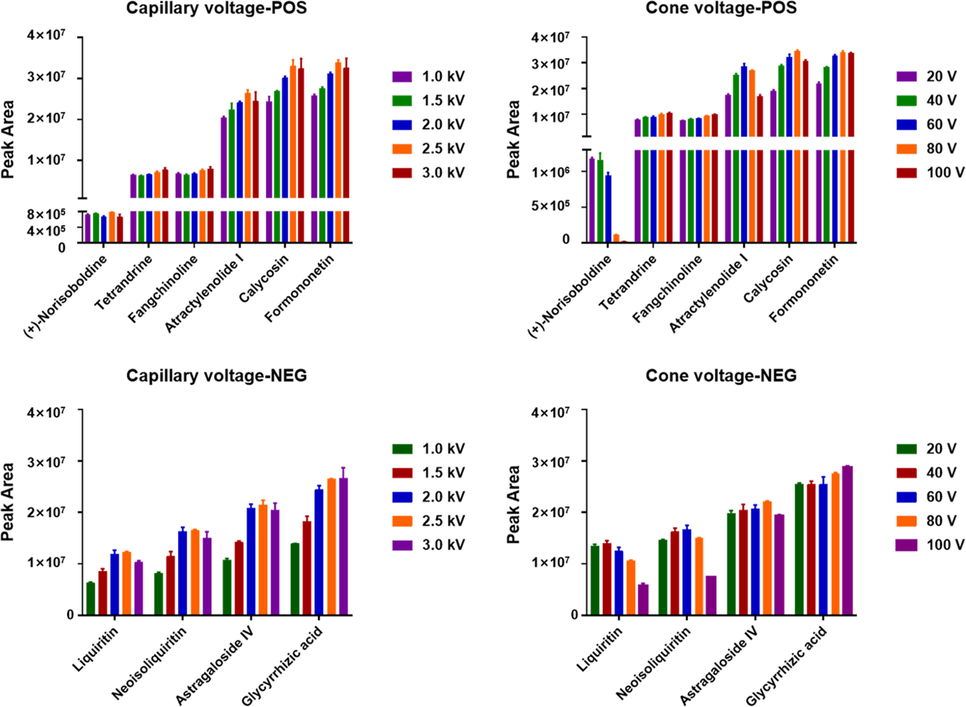
Optimization of the key parameters (capillary voltage and cone voltage) of the Vion IM-QTOF mass spectrometer for the detection of the multicomponents from FHD in both the positive and negative ESI modes (n = 3). The peak areas of the precursor ions of representative compounds are used as the indicators.
3.2 Establishment of PILs for FHD in the setting of HDDDA based on an in-house library and mass defect filtering
Considering the coverage of DDA, even by the HDDDA with IM separation enabled, may be low when facing a complex chemical matrix like the TCM formula FHD, we determined to add PIL in HDDDA in acquiring the CID-MS2 data (Wang et al., 2019), which relied on a self-built database and mass defect filtering. First of all, an in-house library of FHD was established by giving a comprehensive summary of the literature with respect to the phytochemical researches of four component drugs, STR, AR, AMR, and GRR. By searching the public database and reviewing these documents, a total of 730 compounds (including 163 from STR, 179 from AR, 79 from AMR, and 309 from GRR) were included, with their chemical structure, molecular formulae, and source information recorded. An Excel table compatible to the format requirement together with the chemical structures saved as the .mol files were incorporated into the UNIFI software (Wang et al., 2020; Zhang et al., 2019; Zuo et al., 2019). In the next step, a fixed variation range mass defect filtering tool was created to screen the targeted masses from the positive and negative full-scan MS1 data based on the established FHD library. These 730 known compounds were consistent with 406 different masses. In the positive mode, the corresponding protonated form ([M + H]+) and the sodium adduct ([M + Na]+) were considered, generating 809 theoretical m/z values (Fig. 4A), while calculation of the deprotonated form ([M−H]–) and the formic acid adduct ([M−H + FA]–) yielded 805 masses (Fig. 4B). By using the MOD function of Excel, the integer mass and decimal mass of an m/z value were discerned rapidly. The dynamic variation range, {Decimal mass–10 mDa, Decimal mass + 10 mDa}, together with the integer mass, could serve as a “Sieve” to orthogonally screen the target precursor ions from the full-scan raw data. Ultimately, the PILs of FHD in the positive and negative modes involved 316 (Fig. 4C) and 258 (Fig. 4D) m/z values, respectively. The obtained PILs (Table S2 and Table S3) were included in HDDDA to acquire the CID-MS2 data for structural elucidation.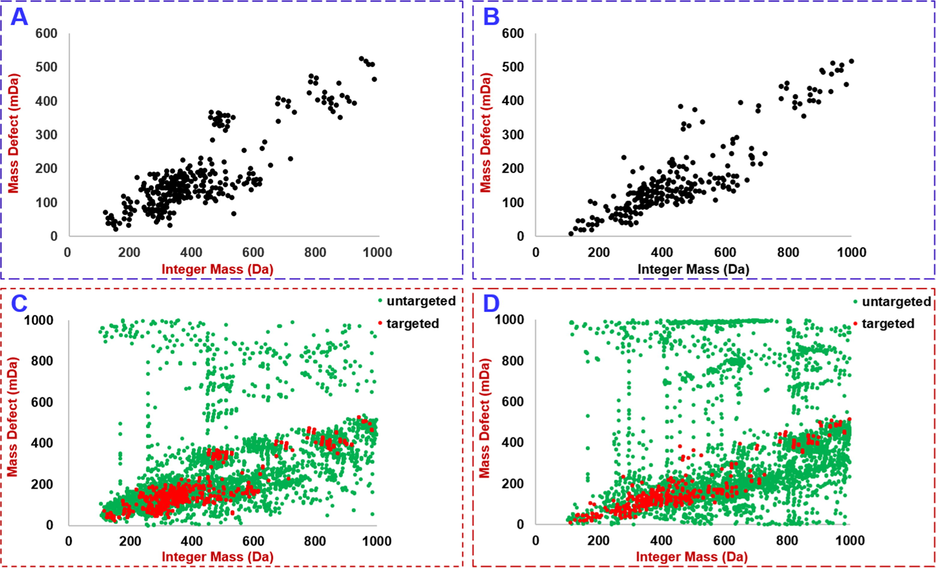
In-house development of a discrete, fixed variation range mass detect filtering tool by plotting the integer mass versus mass defect of 406 masses calculated from 730 known compounds isolated from the four component drugs of FHD (A: positive mode; B: negative mode) and its application to screen the components with the target m/z values from the full-scan MS1 spectra of FHD (C: positive mode; D: negative mode).
To demonstrate the superiority of including PIL in HDDDA (namely PIL/HDDDA) in characterizing the components with those target masses from FHD, the numbers of the compounds that were automatically identified by UNIFI based on the CID-MS2 data acquired by HDDDA and PIL/HDDDA, were compared. By searching against the in-house FHD library, the software was able to match and primarily identify 601 components (including the repeated characterizations in need of the further confirmation process) from 5511 extracted peaks using the PIL/HDDDA data, and 407 components from 5673 extracted peaks using the HDDDA data, in the positive mode. It indicated, by the including of PIL, the identification performance of HDDDA could be improved by 48% in the positive mode. Similarly, in the negative mode, the numbers of the primarily identified components were 421 (totally 5102 peaks) by PIL/HDDDA and 376 (5312 peaks) by HDDDA, which suggested an improvement of 12% in characterizing the target components from FHD in the negative ESI mode. Accordingly to the criteria for selecting the precursor ions to trigger MS/MS fragmentation, by including the PILs, the ions in full-scan MS1 spectra giving the m/z values consistent with those screened target masses (within a predefined variation range) are preferably selected, while no hits are found the mass analyzer operates to perform the fragmentation of those unwanted masses based on the intensity ranking, which renders a simultaneously targeted and untargeted characterization approach similar to the methods we previously established by the Q Exactive Q-Orbitrap mass spectrometer (Fu et al., 2019; Li et al., 2020b; Wang et al., 2019). These data testified the significance of including PIL in DDA, which thus laid a foundation to identify more minor components from FHD.
3.3 Primary comparison of the MS data obtained by HDDDA and the conventional DDA
The influences resulting from the enabling of IM separaton for the established HDDDA approach were evaluated by comparising the MS data acquired on the same Vion IM-QTOF mass spectrometer using the conventional DDA method. First, the ion response for the precusor ions of seven selected index components (m/z 609.2970 for fangchinoline, m/z 623.3126 for tertrandrine, and m/z 231.1384 for atractylenolide I detected in ESI+; m/z 549.1604 for liquiritin apioside, m/z 417.1178 for liquiritin isomer, m/z 829.3808 for astragaloise A, and m/z 821.3988 for glycyrrhizic acid in ESI− ) was less intense in HDDDA in contrast to DDA (Table S4), which may be attributed to the additional IM separation facilitated by HDDDA. The weaker ion response also significantly decreased the signal-to-noise (S/N) ratios in HDDDA. Second, the MS1 spectra obtained by HDDDA for four compounds (fangchinoline, atractylenolide I, liquiritin apioside, and glycyrrhizic acid) became much more cleaner (dubbed high difinition), as those co-eluting component could be further resolved by the IM separation (Figure S1). Less interefence in the MS spectra could benefit the accurate matching by the UNIFI software. Third, the CCS value for each compound could be conveniently determined by the HDDDA approach, and the significant difference in CCS might be of the potential to discriminate the isomeric compounds as listed in Table S5. The features mentioned above can demonstrate the good performance of the established HDDDA approach supporting the comprehensive multicomponent characterization of FHD.
3.4 Comprehensive multicomponent characterization of FHD by analyzing the negative and positive CID-MS2 data obtained by the established HDDDA approaches
Due to the enabling of IM separation using the Vion IM-QTOF mass spectrometer, an HDDDA approach was established, by which high-definition MS2 data could be acquired, and simultaneously, the drift time or CCS of each component was conveniently determined. Our recent research has demonstrated the additional significance of IM separation, aside from the ion selection by quadrupole (Q), in more accurate analysis of the target components (Zhang et al., 2020). Fig. 5 shows the base peak intensity (BPI) chromatograms of FHD recorded in both the positive and negative ESI modes. To streamline the peak annotation of the high-resolution MS2 data recorded by UNIFI, standardized workflows were established similar to those utilized in processing the HDMSE (high-definition MSE, a data-independent acquistion approach) data (Koley et al., 2020; Zhang et al., 2019; Zuo et al., 2019). Matching by UNIFI was performed between the experimental CID-MS2 data and the predicted data (MS1 and MS2) based on the in-house library within the pre-defined mass error range. Based on the matching results listed as “Identified Components” (those conforming to the matching rules) and “Unknown Components” (those unconforming to the matching rules), the subsequent re-confirming process was necessary to remove the false positives and check the identities by analyzing the fragmentation pathways and the retention rules. Consequently, we were able to identify or tentatively characterize totally 203 components from FHD, including the alkaloids, lactones, flavonoids, and triterpenoids/saponins, etc.), with the information detailed in Table S6. Amongst them, alkaloids and lactones were characterized based on the positive CID-MS2 data, while the other catogeries were based on the negative data. To sum up, the neutral loss masses of the sugars eliminated from the deprotonated precursors or the FA-adduct molecules included 162.05 Da for glucose (Glc), 146.06 Da for rhamnose (Rha), 176.03 Da for glucuronic acid (GlurA), and 132.04 Da for both the xylose (Xyl) and arabinose (Ara). For a convenient description of the characterized glycosides, we used Xyl to represent all the possible pentoses characterized by the neutral loss of 132 Da, in this work.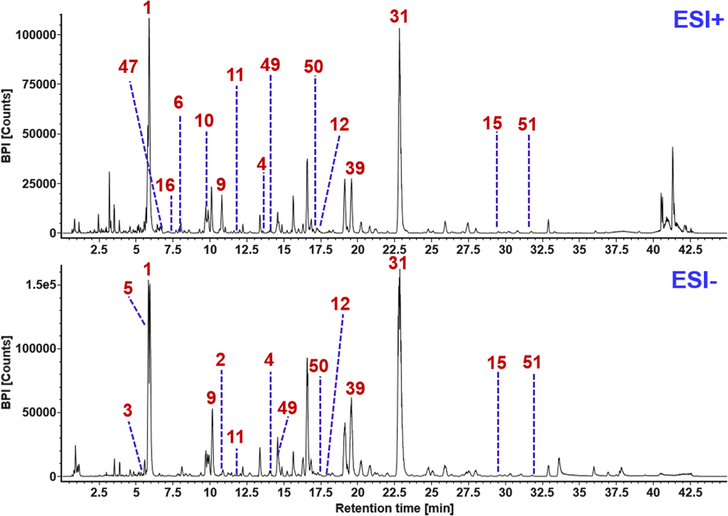
Base peak intensity chromatograms of FHD in both the positive and negative ESI modes. The peaks identified by reference compounds comparison are annotated with the numbers consistent with the order in Fig. 1.
3.4.1 Characterization of the alkaloids
Alkaloids are nitrogen-containing alkaline organic compounds widely distributed in plants, which have been reported with significant biological activities (Chen et al., 2020). Alkaloids detected in FHD by the positive ESI mode were mostly ascribed to STR, which has been reported to contain six subtypes of alkaloids: aporphine, morphine, bisbenzylisoquinoline, tetrahydrotrotoberberine/berberine, benzylisoquinoline, and nitrophenanthrene carboxylic acid, etc. (Sim et al., 2015). In this work, we were able to characterize 25 alkaloids. Their main CID-MS2 fragmentation pathways involved the eliminations of methylamine (NH2CH2), methyl (•CH3), methanol (CH3OH), CO, and RDA cleavages, etc., which could be partially reflected by the CID-MS2 spectrum of a bisbenzylisoquinoline-type alkaloid reference compound, tetrandrine (consistent with compound 76# in Table S6: tR 9.35 min, C38H42N2O6) as illustrated in Fig. 6. It gave an obvious protonated precursor ion ([M + H]+) at m/z 623.3116, and upon CID-MS2 dissociation, three rich product ions were obtained: the fragment at m/z 592.2706 should result from the loss of NH2CH3 (31.04 Da); the ion at m/z 381.1800 was a fragment due to the simultaneous elimination of rings C and D together with two methylene groups (C2H4); and the product ion m/z 192.1023 was most likely to be a single isoquinoline moiety. These data generally accorded with those reported in literature (Sim et al., 2015). There regular fragmentation behaviors could provide diagnostic information to identify those unknown alkaloids detected from FHD, such as compound 7# (tR 3.32 min). Its MS1 spectrum showed an abundant parent ion at m/z 300.1592, which suggested the molecular formula C18H21NO3 (mass error: –0.9 ppm). Similar to compound 76#, the transition m/z 300.01 → 255.10 was consistent with the elimination of NH(CH3)2 or NH2CH3 + CH2 (45.06 Da), while the product ion m/z 237.0921 should be the secondary fragment of m/z 255.10 by further dissociating a molecule of H2O. A low-mass fragment m/z 107.0476 should be ascribed to the benzyl moiety (C7H7O+), a common fragment for the benzylisoquinoline alkaloids of STR. By comparing the fingerprints of four single drugs, it was confirmed to be from STR. Based on these evidences and by referring to the in-house library of FHD, compound 7# was tentatively characterized as N-methylcoclaurine (Jiang et al., 2020). Information of the other 23 alkaloid compounds that have been characterized from FHD is given in Table S6.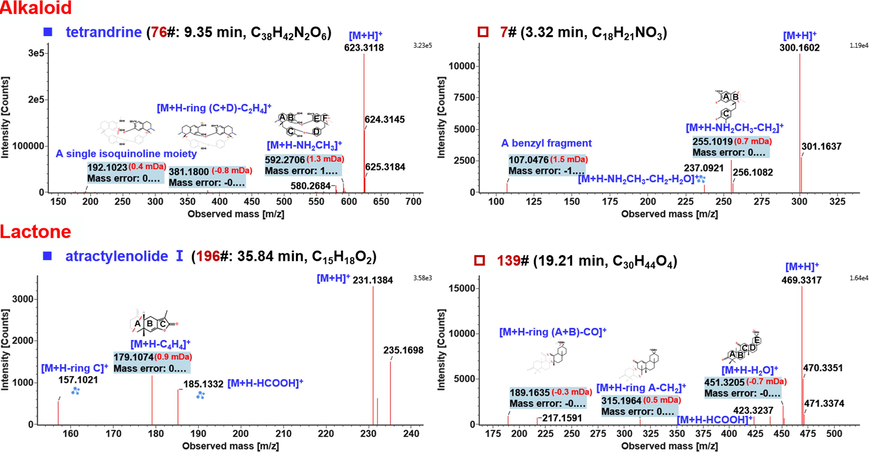
Annotation of the CID-MS2 spectra of the representative compounds for the alkaloids (compounds 76# and 7#) and lactones (compounds 196# and 139#), identified from FHD based on the positive CID-MS2 data.
3.4.2 Characterization of the lactones
Lactones are a class of low-polarity biologically active ingredients with ring-infused ester bond. The lactones detected from FHD are mainly derived from AMR (atractylenolides, belonging to the sesquiterpenoid) and a small proportion as triterpene lactones from GRR (Ji et al., 2014; Qian et al., 2021; Sun et al., 2017). In this research, we identified 16 lactones, for which a characteristic cleavage was the neutral loss of HCOOH closely related to the presence of a lactone structure. In this section, the structural elucidation of compounds 196# (tR 35.84 min, C15H18O2) and 139# (tR 19.21 min, C30H44O4) was intepreted (Fig. 6). Compound 196# was identified as atractylenolide I with the aid of the reference compound comparison. The protonated precursor ion was observed at m/z 231.1378, the CID-MS2 of which could yield the fragment of m/z 185.1332 by the neutral elimination of HCOOH (46.01 Da) with the lactone ring cleaved. Another fragment at m/z 179.1074 should result from the partial cleavage of ring A with C4H4 lost. The third product ion at m/z 157.1021 was assigned as a fragment due to the elimination of the whole ring C. Another lactone compound was a lactone of tetracyclic triterpene (compound 139#) with the protonated precursor ion observed at m/z 469.3315 (mass error: 0.6 ppm). Upon CID, the characteristic fragmentation by neutral loss of HCOOH generated a product ion at m/z 423.3237. Moreover, the neutral elimination of H2O yielded a product ion of m/z 451.3205. The other two fragments at m/z 315.1964 and 189.1635 could be ascribed to the ring-opening products. After comparing with the retention behavior of the single drugs and referring to the UNIFI identification results, we tentatively characterized compound 139# as uralenolide, a triterpenoid lactone from GRR (Sun et al., 1987).
3.4.3 Characterization of the flavonoids
Flavonoids refer to a series of widely distributed natural compounds bearing a “C6-C3-C6” skeleton, which can also be considered to derive from 2-phenylchromone. We were able to characterize a total of 86 flavonoid compounds from FHD based on the negative-mode CID-MS2 data obtained by HDDDA, (occupying 42.36% of the total amount), which are known as one class of main active ingredients in AR and GRR (Guo et al., 2019; Xiang et al., 2012). The negative-mode fragmentation features of flavonoids mainly included a series of neutral eliminations of CO2, CO, the radical cleavage of •CH3 (for those containing OCH3), and RDA dissociation, etc. (Yang et al., 2012). The CID-MS2 features of an isoflavonoid reference compound, formononetin (corresponding to compound 130#: tR 16.96 min, C16H12O4), were illustrated (Fig. 6). Evidently, the deprotonated precursor ion at m/z 267.0663, upon CID, was prone to eliminate •CH3 and CH4 generating the product ions of m/z 252.0426 and 251.0340, respectively. These two accompanying ions could further eliminate CO and 2 × CO, forming the fragments of m/z 224.0440/223.0400 and 196.0493/195.0451, respectively. What’s more, the ion of m/z 167.0499 should result from the loss of 3 × CO from the CH4-lost fragment m/z 251. The fragment with the lowest mass at m/z 132.0213 was a product ion of ring B with the cleavage occurring to ring C. Compound 104# (tR 13.25 min) gave an abundant precursor ion at m/z 283.0612 (0 ppm), suggesting the molecular formula C16H12O5 with one O atom more than compound 130#. Similar to formononetin, the CID-MS2 of compound 104# gave the product ions by the elimination of •CH3 and CH4 (m/z 267.0289), and the combinational loss of CO (m/z 239.0349), 2 × CO (m/z 211.0400), and even 3 × CO (m/z 183.0448). In addition, two weak fragments at m/z 135.0087 (1.4A-) and 119.0135 (0.4A-) may be the RDA fragments, which could indicate the presence of OH and OCH3 on ring A. Based on these evidences and by referring to the FHD library, compound 104# was tentatively characterized as prunetin, a bioactive isoflavone from GRR (Ahu et al., 2013).
3.4.4 Characterization of the triterpenes and saponins
Triterpenes and the glycosides (saponins) are another type of main bioactive ingredients in AR and GRR (Guo et al., 2019; Xiang et al., 2012). A total of 48 triterpenes/saponins were characterized from FHD in this study. The regular fragmentation pathways of these saponins involved the neutral loss of the sugar moieties, the generation of the deprotonated sapogenin ions, and even the fragments of the oligosaccharides (Ji et al., 2014). These features embodied by the negative CID-MS2 could be evident from the spectrum of a saponin reference compound, glycyrrhizic acid (164#: tR 24.63 min, C46H62O16) as illustrated in Fig. 7. The negative-mode CID-MS2 of the deprotonated precursor of m/z 821.3963 could sequentially eliminate GlurA (m/z 645.3650) and 2 × GlurA (m/z 469.3316), generating a medium intensity of sapogenin ion at m/z 469.3316. Moreover, the fragments of m/z 627.3536 and 583.3635 were reasonably assigned as [M−H−GlurA−H2O]– and [M−H−GlurA−H2O−CO2]–, respectively. In addition, due to the strong electronegativity, the deprotonated GlurA-GlurA moiety could generate a predominant fragment at m/z 351.0568, which could be diagnostic to characterize the oligosaccharide GlurA-GlurA in saponins from GRR. Moreover, two secondary fragments of the GlurA-GlurA were observed at m/z 289.0569 and 193.0352, which were ascribed to [GlurAGlurA–H–CO2–H2O]– and [GlurA + H2O–H]–, respectively. Based on these fragmentation features, we illustrated the characterization of an unknown saponin, compound 126# (tR 16.52 min). It gave a rich deprotonated precursor ion at m/z 983.4485 (mass error: –0.8 ppm), the CID-MS2 of which could generate a series of the product ions by the neutral elimination of sugars and the generation of the sapogenin product ion, as shown in Fig. 7. These product ions were almost the same as those obtained for compound 164# beginning with the fragment by losing Glc (m/z 821.3961). These data was suggestive of an additional Glc in this structure compared with compound 164#. After matching with the database and the retention behavior of the single medicine, we could tentatively characterize compound 126# as licorice-saponin A3 (Montero et al., 2016). The saponins identified from AR showed similar fragmentation pathways, and the neutrally lost masses and sapogenin ions were diagnostic for their tentative characterization.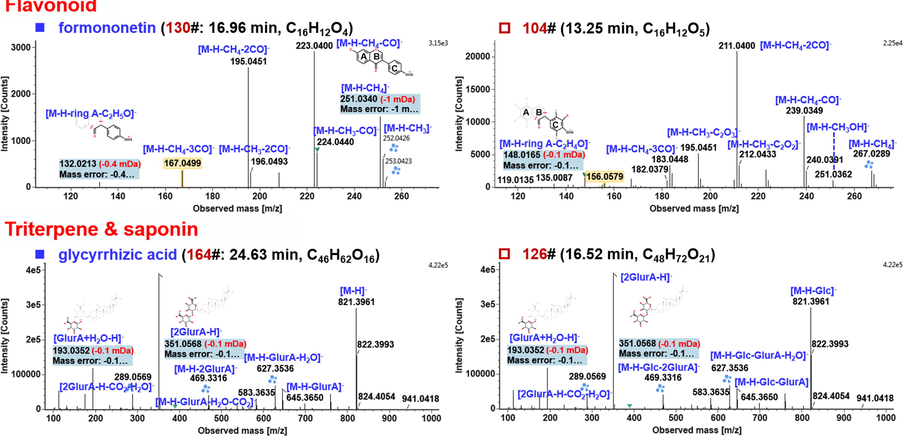
Annotation of the CID-MS2 spectra of the representative compounds for the flavonoids (compounds 130# and 104#) and triterpenes/saponins (compounds 164# and 126#), identified from FHD based on the negative CID-MS2 data.
In addition to these four types of herbal components, a total of 28 compounds classified as the others were characterized from FHD as well, with their information detailed in Table S6.
Despite the merits depicted above, there’s still some insufficiency for the established UHPLC/IM-QTOF-HDDDA approach. On one hand, the determined CCS values have not been fully utilized in confirming the structural identification results. On the other hand, numerous unknown components have been found, but we fail to identify their structures only based on the limited MS information, which may represent novel substances in the FHD formula we investigate. It becomes urgent to elaborate the extensive CCS library for natural products, and we shall endeavor to contribute to this filed as our future work.
4 Conclusions
With the view of in-depth deconvolution of the chemical complexity of the TCM formulae, this work, using FHD as a case, presented an IM-enabled and PIL-included UHPLC/IM-QTOF-HDDDA approach. Good chromatographic separation of the multicomponents from FHD was achieved by configuring a CORTECS UPLC T3 column and utilizing 0.1% FA in H2O/0.1% FA in CH3CN as the mobile phase. Key parameters in method development of HDDDA were carefully optimized in pursuit of the best performance, in particular, the inclusion of PILs in both the positive and negative ESI modes could largely improve the coverage of those target components. The enabling of IM separation could generate high-definition MS spectra and conveniently determine the CCS values, the potential role of which in structural identification deserved further research. The in-house library-driven automatic peak annotation by UNIFI was proven to be a practical tool to efficiently process the HDDDA data in support of the comprehensive multicomponent characterization of TCM or the comprehensive metabolites identification. By these efforts, we were able to identify or tentatively characterize 203 components from FHD, which indicated approximately 2.36 folds (203 versus 86) of the compounds ever-reported in literature (Yang et al., 2020). To our knowledge, it is the first report regarding the method development of HDDDA that targets the global TCM components characterization, and the data obtained are beneficial to the quality control and the secondary development of FHD.
Acknowledgments
This work was financially supported by National Key R&D Program of China (Grant No. 2018YFC1704500), National Natural Science Foundation of China (Grant No. 81872996), Tianjin Municipal Education Commission Research Project (Grant No. 2017ZD07), and Scientific Research project in the Key Field of Traditional Chinese medicine of Tianjin Municipal Health Commission (Grant No. 2021005).
Declaration of Competing Interest
The authors declared that there is no conflict of interest.
References
- Molecular mechanisms underlying the anti-obesity potential of prunetin, an O-methylated isoflavone. Biochem. Pharmacol.. 2013;85(10):1525-1533.
- [CrossRef] [Google Scholar]
- Lycopodium japonicum: A comprehensive review on its phytochemicals and biological activities. Arab. J. Chem.. 2020;13(5):5438-5450.
- [CrossRef] [Google Scholar]
- Simultaneously targeted and untargeted multicomponent characterization of Erzhi Pill by offline two-dimensional liquid chromatography/quadrupole-Orbitrap mass spectrometry. J. Chromatogr. A. 2019;1584:87-96.
- [CrossRef] [Google Scholar]
- A holistic approach to the quality control of traditional Chinese medicines. Science. 2015;347:S29-S31.
- [Google Scholar]
- Comprehensive characterization of lignans from Forsythia viridissima by UHPLC-ESI-QTOF-MS and their NO inhibitory effects on RAW 264.7 cells. Molecules. 2019;24(14):2649.
- [CrossRef] [Google Scholar]
- New triterpene saponins from the roots of Glycyrrhiza yunnanensis and their rapid screening by LC/MS/MS. J. Pharm. Biomed. Anal.. 2014;90:15-26.
- [CrossRef] [Google Scholar]
- A critical review: traditional uses, phytochemistry, pharmacology and toxicology of Stephania tetrandra S. Moore (Fen Fang Ji) Phytochem. Rev.. 2020;19(2):449-489.
- [CrossRef] [Google Scholar]
- High resolution LC-MS characterization of phenolic compounds and the evaluation of antioxidant properties of a tropical purple radish genotype. Arab. J. Chem.. 2020;13(1):1355-1366.
- [CrossRef] [Google Scholar]
- A modified data filtering strategy for targeted characterization of polymers in complex matrixes using drift tube ion mobility-mass spectrometry: Application to analysis of procyanidins in the grape seed extracts. Food Chem.. 2020;321:126693.
- [CrossRef] [Google Scholar]
- MetDIA: Targeted metabolite extraction of multiplexed MS/MS spectra generated by data-independent acquisition. Anal. Chem.. 2016;88(17):8757-8764.
- [CrossRef] [Google Scholar]
- Integration of multicomponent characterization, untargeted metabolomics and mass spectrometry imaging to unveil the holistic chemical transformations and key markers associated with wine steaming of Ligustri Lucidi Fructus. J. Chromatogr. A. 2020;1624:461228.
- [CrossRef] [Google Scholar]
- Volatile component interaction effects on compatibility of Cyperi Rhizoma and Angelicae Sinensis Radix or Chuanxiong Rhizoma by UPLC-MS/MS and response surface analysis. J. Pharm. Biomed. Anal.. 2018;160:135-143.
- [CrossRef] [Google Scholar]
- A data-independent acquisition workflow for qualitative screening of new psychoactive substances in biological samples. Anal. Bioanal. Chem.. 2015;407(29):8773-8785.
- [CrossRef] [Google Scholar]
- Metabolite profiling of licorice (Glycyrrhiza glabra) from different locations using comprehensive two-dimensional liquid chromatography coupled to diode array and tandem mass spectrometry detection. Anal. Chim. Acta. 2016;913:145-159.
- [CrossRef] [Google Scholar]
- Metabolomic analysis of the effects of cadmium and copper treatment in Oryza sativa L. using untargeted liquid chromatography coupled to high resolution mass spectrometry and all-ion fragmentation. Metabolomics. 2017;9(6):660-675.
- [CrossRef] [Google Scholar]
- Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal. Chem.. 2015;87(2):1137-1144.
- [CrossRef] [Google Scholar]
- An integrated approach for global profiling of multi-type constituents: Comprehensive chemical characterization of Lonicerae Japonicae Flos as a case study. J. Chromatogr. A. 2020;1613:460674.
- [CrossRef] [Google Scholar]
- A four-dimensional separation approach by offline 2D-LC/IM-TOF-MS in combination with database-driven computational peak annotation facilitating the in-depth characterization of the multicomponents from Atractylodis Macrocephalae Rhizoma (Atractylodes macrocephala) Arab. J. Chem.. 2021;14(2):102957.
- [CrossRef] [Google Scholar]
- A green protocol for efficient discovery of novel natural compounds: characterization of new ginsenosides from the stem and leaves of Panax ginseng. Anal. Chim. Acta. 2015;893:65-76.
- [CrossRef] [Google Scholar]
- Metabolite identification using an ion mobility enhanced data-independent acquisition strategy and automated data processing. Rapid Commun. Mass Spectrom.. 2020;34(12) e8792
- [CrossRef] [Google Scholar]
- Two-dimensional liquid chromatographic analysis of ramelteon in human serum. Arab. J. Chem.. 2019;12(8):2817-2822.
- [CrossRef] [Google Scholar]
- Identification of alkaloid constituents from Fangchi species using pH control liquid-liquid extraction and liquid chromatography coupled to quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom.. 2015;29(9):837-854.
- [CrossRef] [Google Scholar]
- A classification of liquid chromatography mass spectrometry techniques for evaluation of chemical composition and quality control of traditional medicines. J. Chromatogr. A. 2020;1609:460501.
- [CrossRef] [Google Scholar]
- Influence of sulfur fumigation on the chemical profiles of Atractylodes macrocephala Koidz. evaluated by UFLC–QTOF–MS combined with multivariate statistical analysis. J. Pharm. Biomed. Anal.. 2017;141:19-31.
- [CrossRef] [Google Scholar]
- Isolation and structure determination of new triterpene sapogenins from Glycyrrhiza uralensis Fisch. Acta Pharm. Sin.. 1987;22(7):512-514.
- [CrossRef] [Google Scholar]
- MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods. 2015;12(6):523-526.
- [CrossRef] [Google Scholar]
- The emerging of ion mobility-mass spectrometry in lipidomics to facilitate lipid separation and identification. Trends Anal. Chem.. 2019;116:332-339.
- [CrossRef] [Google Scholar]
- Ultra high performance liquid chromatography with tandem mass spectrometry method for the determination of tetrandrine and fangchinoline in rat plasma after oral administration of Fangji Huangqi Tang and Stephania tetrandra S. Moore extracts. J. Sep. Sci.. 2015;38(8):1286-1293.
- [CrossRef] [Google Scholar]
- Research progress in chemical constituents in single herbs of Fangji Huangqi Decoction and its pharmacological activities. Chin. Tradit. Herbal Drugs. 2016;47(19):3527-3534.
- [CrossRef] [Google Scholar]
- Screening and identification of multiple constituents and their metabolites of Fangji Huangqi Tang in rats by ultra-high performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry basing on coupling data processing techniques. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci.. 2015;985:14-28.
- [CrossRef] [Google Scholar]
- Development and validation of an UHPLC-QqQ-MS technique for simultaneous determination of ten bioactive components in Fangji Huangqi Tang. J. Anal. Methods Chem.. 2016;2016:1-8.
- [CrossRef] [Google Scholar]
- Holistic quality evaluation of Saposhnikoviae Radix (Saposhnikovia divaricata) by reversed-phase ultra-high performance liquid chromatography and hydrophilic interaction chromatography coupled with ion mobility quadrupole time-of-flight mass spectrometry-based untargeted metabolomics. Arab. J. Chem.. 2020;13(12):8835-8847.
- [CrossRef] [Google Scholar]
- In-depth profiling, characterization, and comparison of the ginsenosides among three different parts (the root, stem leaf, and flower bud) of Panax quinquefolius L. by ultra-high performance liquid chromatography/quadrupole-Orbitrap mass spectrometry. Anal. Bioanal. Chem.. 2019;411(29):7817-7829.
- [CrossRef] [Google Scholar]
- Rapid screening and characterization of triterpene saponins in Acanthopanax senticosus leaves via untargeted MSAll and SWATH techniques on a quadrupole time of flight mass spectrometry. J. Pharm. Biomed. Anal.. 2019;170:68-82.
- [CrossRef] [Google Scholar]
- Collision-induced dissociation of 40 flavonoid aglycones and differentiation of the common flavonoid subtypes using electrospray ionization ion-trap tandem mass spectrometry and quadrupole time-of-flight mass spectrometry. Eur. J. Mass Spectrom.. 2012;18(6):493-503.
- [CrossRef] [Google Scholar]
- Classification and distribution analysis of components in Glycyrrhiza using licorice compounds database. Acta Pharm. Sin.. 2012;47(8):1023-1030.
- [CrossRef] [Google Scholar]
- Systematically characterize the absorbed effective substances of wutou decoction and their metabolic pathways in rat plasma using UHPLC-Q-TOF-MS combined with a target network pharmacological analysis. J. Pharm. Biomed. Anal.. 2017;141:95-107.
- [CrossRef] [Google Scholar]
- Simultaneous determination of components with wide polarity and content ranges in Cistanche tubulosa using serially coupled reverse phase-hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A. 2017;1501:39-50.
- [CrossRef] [Google Scholar]
- Comprehensive investigation of mechanism and effective ingredients of Fangji Huangqi Tang by serum pharmacochemistry and network pharmacology. Biomed. Chromatogr.. 2020;34(4) e4785
- [CrossRef] [Google Scholar]
- DecoMetDIA: deconvolution of multiplexed MS/MS spectra for metabolite identification in SWATH-MS based untargeted metabolomics. Anal. Chem.. 2019;91(18):11897-11904.
- [CrossRef] [Google Scholar]
- Highly selective monitoring of in-source fragmentation sapogenin product ions in positive mode enabling group-target ginsenosides profiling and simultaneous identification of seven Panax herbal medicines. J. Chromatogr. A. 2020;1618:460850.
- [CrossRef] [Google Scholar]
- Integration of data-dependent acquisition (DDA) and data-independent high-definition MSE (HDMSE) for the comprehensive profiling and characterization of multicomponents from Panax japonicus by UHPLC/IM-QTOF-MS. Molecules. 2019;24(15):2708.
- [CrossRef] [Google Scholar]
- Recent pharmaceutical evidence on the compatibility rationality of traditional Chinese medicine. J. Ethnopharmacol.. 2017;206:363-375.
- [CrossRef] [Google Scholar]
- Application of two-dimensional liquid chromatography in the separation of traditional Chinese medicine. J. Sep. Sci.. 2020;43(1):87-104.
- [CrossRef] [Google Scholar]
- Large-scale prediction of collision cross-section values for metabolites in ion mobility-mass spectrometry. Anal. Chem.. 2016;88(22):11084-11091.
- [CrossRef] [Google Scholar]
- LipidCCS: prediction of collision cross-section values for lipids with high precision to support ion mobility-mass spectrometry-based lipidomics. Anal. Chem.. 2017;89(17):9559-9566.
- [CrossRef] [Google Scholar]
- Data-dependent acquisition and database-driven efficient peak annotation for the comprehensive profiling and characterization of the multicomponents from compound Xueshuantong capsule by UHPLC/IM-QTOF-MS. Molecules. 2019;24(19):3431.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2021.103087.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







