Application of G-quadruplex aptamer conjugated MSNs to deliver ampicillin for suppressing S. aureus biofilm on mice bone
⁎Corresponding author. h.mohabatkar@ast.ui.ac.ir (Hassan Mohabatkar)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Staphylococcus aureus is a common bacterial agent of biofilm formation in medical environments. The formed biofilm of this bacterium in bone tissue is one of the main causes of osteomyelitis, which is a serious health issue. Due to the importance of this infection after traumatic injuries or surgical intervention, it is necessary to develop a system that could release the antibiotics at the site of injury, specifically and gradually. The current study aimed to develop a nanosystem composed of single-stranded G-quadreplex DNA aptamer as the bio-recognition element, mesoporous silica nanoparticles (MSNs) as the carrier for gradual drug release, and Ampicillin as the cargo to be delivered to the site of infection. In silico methods were used to select an optimum binding aptamer against protein A of S. aureus. The binding of aptamer was confirmed via gel retardation assay, DLS, and Zeta potential analyses. The loading of the drug was confirmed by the FTIR method, and the drug release investigation showed almost 30 % of drug release via 48 h dialysis assay. The acquired results from the biofilm suppression assay indicated that this system provides a significant inhibitory effect against the S. aureus biofilm and has a high potential for the desired drug release to prevent the formation of biofilm, and could destroy the biofilm on the mice bone. The results of the MTT assay proved that this system does not pose a significant toxicity thread for MCF-7 cell viability, as a model for eukaryotic cells. In vivo studies are required to further confirm the efficacy of this system against S. aureus biofilm on bone.
Keywords
Biofilm inhibition
Mesoporous silica nanoparticles
G-quadreplex DNA aptamers
Drug delivery
Data availability statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
1 Introduction
Staphylococcus aureus is an opportunist pathogen that is available on the surface of the human body skin, and inside of cavities such as the mouth or throat. This microorganism usually does not cause any serious problem, as long as the human body and immune system are in normal condition. Although, when this pathogen finds any opportunity, for example at the time of surgeries or in open fractures, it exacerbates the damage conditions (Raineri et al., 2021; González-García et al., 2021; Zhang, 2021). The biofilm of this bacteria is one of the most common causes of morbidity in various infections that could be formed alone or along with other pathogens, such as Streptococal and Staphylococcal ones (Gupta, 2021; Barraza and Whiteley, 2021).
One of the most serious infections related to the biofilm of S. aureus is osteomyelitis (Gao, 2021). Osteomyelitis is defined as the infection in bone tissue that mostly happens after an open fracture caused by trauma or surgical interventions (Van Vugt, 2021). This condition is one of the disorders requiring immediate medical care. Otherwise, it could end up in dramatic consequences, such as long-term morbidity, and amputation requirement, and in some cases, it could even cause septicemia that may result in losing the patient’s life (Wu, 2020; Bessesen, 2020; Nasser, 2020).
As mentioned above, surgical intervention is one of the risk factors for causing surgical site infection (SSI) and perhaps osteomyelitis (Watanabe, 2010). Orthopedic surgeries are categorized as intensive operations, and sometimes the bone fixation might require extra time, which elevates the risk for infection with opportunist pathogens such as S. aureus. For this reason, in some orthopedic surgeries, such as those involving the installation of implants, surgeons use local antibiotic therapy and surgical pads for the gradual drug release at the site of infection (Masters, 2019).
Nanotechnology has revolutionized the modern medical and biological studies by providing tools such as drug delivery and regenerative techniques Kazemi et al., 2018. Targeted drug delivery is one of the novel phenomena in modern medicine that has the potential for increasing the efficacy of regular therapeutics (Manzari, 2021). In this regard, the science of nanotechnology is helpful for researchers in this area by providing various nanoparticles that could be acting as therapeutics themselves, or provide carriers for the drugs; such as anticancer agents (Gharbavi, 2021) or antibiotics (Baranyai, 2021). The emergence of core-shell nanoparticles, noisosomes, and theranostic nanoparticles have shown their efficacy in specific targeted delivery of medical agents against various disorders (Gharbavi, 2020; Nosrati, 2022).
One of the novel systems for drug delivery is the application of a three-component system, which is firstly has of a bio-recognition element that could be either a molecular aptamer or a monoclonal antibody. The second component is a carrier, which acts as a storage for the selected therapeutic agent. In this regard, mesoporous silica nanoparticles (MSNs) are one of the popular carriers. Due to the high ratio of surface area to volume, MSNs could absorb a high load of the drug as cargo, and release it gradually into the desired site. The third part of this three-component system is the selected therapeutic agent that is loaded inside of the carrier and released into the target environment (Ozalp et al., 2011; Xie, 2016).
Nucleic acid aptamers are single-stranded sequences of oligonucleotides that form a unique tertiary structure, which is capable of specific binding to a molecular target (Ni, 2011). These structures have attracted scientific attention toward themselves as bio-recognition elements, due to their specificity, easy synthesis, and lower production price, compared to monoclonal antibodies (Liu, 2021). The base structure for such sequences is usually provided through an in vitro selection of the most specific sequences to bind a target, followed by an increase and selection of the ones with higher affinity, which is called SELEX (Systematic Evolution of Ligands by Exponential Enrichment) method (Lyu and Khan, 2021). However, many studies have reported that it is possible to make mutations in the sequences of DNA aptamers and present new aptamers with higher efficacy and specificity (Navien, 2021).
One of the recent advancements that has revolutionized modern medicine is the emergence of bioinformatics (Behbahani, 2020). The application of in silico tools has reduced the pressure of using the laboratory workforce and has diminished the waste of laboratory materials by providing predictions about biological systems (Mohabatkar et al., 2021). These tools come helpful for a variety of applications; such as designing vaccines (Gharbavi, 2021) or predicting the structure of unknown structures; including novel single-stranded DNA aptamers (Navien, 2021). Moreover, the advent of techniques; such as molecular docking for the prediction of the potential receptor and ligands interactions has shown to be presenting accurate predictions (Haghighi and Moradi, 2020). Previously, these in silico techniques have been reported to provide high-quality predictions for potential interactions between oligonucleotide ligands and protein receptors (Gharbavi, 2020; Johari, 2020).
At the current study, it was aimed to develop a three-component system, composed of a novel G-quadreplex ssDNA aptamer to target protein A of S. aureus as the bio-recognition element to specifically target the biofilm, MSNs as carriers for the selected anti-bacterial agent, and Ampicillin as the cargo to suppress the bacterial biofilm. Mice were used as the animal model, and bone tissues from the skull were used for biofilm formation. Fig. 1 provides a brief outlook on the procedure and outcome of using such a system on bone biofilm induced by S. aureus.
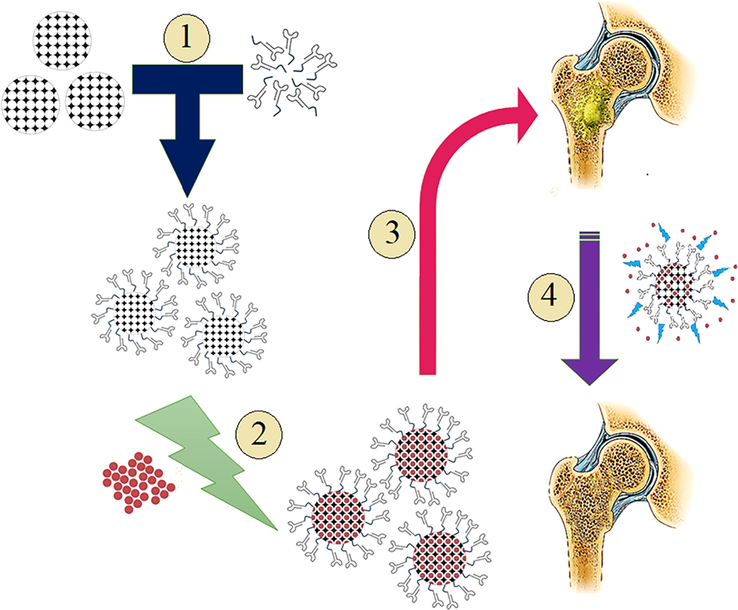
- Schematic workflow of using nanosystem of MSNs- Aptamer-Ampicillin to suppress S. aureus biofilm formation in bone: 1. Conjugation of aptamer and MSNs via disulfide binding, 2. Loading of Ampicillin antibiotic inside of the MSN-aptamer conjugates, 3. Introducing nanosystem to the bone biofilm, 4. Gradual drug release from the system suppresses the formation of S. aureus biofilm.
2 Materials and methods
2.1 Data collection
The required information about the protein A of S. aureus was collected from the protein data bank (PDB). Since protein A is a homo-domain, domain C of this protein (PDB ID: 4NPE) was used as the receptor for the aptamer candidates.
In the case of aptamers, the initial sequence of a specific G-quadruplex aptamer (that was selected by the SELEX assay) was fetched from the literature. The original sequences of the aptamer were reported by Stoltenburg et al. (Stoltenburg et al., 2015) as (PA 76): 3’ATACCAGCTTATTCAATTAGCAACATGAGGGGGATAGAGGGGGTGGGTTCTCTCGGCTACAATCGTAATCAGTTAG 5’. Another study by the same authors reported that the truncated form of these aptamers could effectively bind to protein A, and did not react with related proteins such as Protein G and Protein L. They reported that 5’ site bases are necessary for the formation of the G-quadreplex form, and no base change was allowed in the 3’ATACCAGCTTATTCAATT 5’ nucleotides (Stoltenburg, 2016).
2.2 In silico selection of the most sensitive aptamer and aptamer synthesis
Molecular docking between various DNA constructs and the target protein receptors has shown to be an effective method (Behbahani et al., 2021). To select the most effective G-quadreplex aptamer to target protein A, which is a main component in the S. aureus biofilm, the first step was to provide a 3D conformation of the proposed aptamers. To provide these structures, the in silico method proposed by Heiat et al. (2016) was followed, which has been reported efficient by other studies (Yarizadeh, 2019; Jia, 2020; Presnell and Alper, 2018; Wang, 2019).
The secondary structure of the single-stranded DNA aptamers was predicted using the Mfold web server (version 3.1, online: http://unafold.rna.albany.edu) (Zuker, 2003). Vienna output format sequences (dot-bracket notation) were used for the construction of 3-D structure models of the aptamer.
Molecular docking analysis was carried out using the HADDOCK tool (De Vries et al., 2010) (Version 2.4 available at https://wenmr.science.uu.nl/haddock2.4/), which is appropriate for analyzing the molecular interactions between proteins and DNAs. The optimized structures of the proteins and the aptamers were used as the receptor and ligands in the PDB format.
The initially predicted models, which presented the highest socking scores, were selected for analyzing the interactions. Protein-Ligand Interaction Profiler (PLIP) tool (https://projects.biotec.tu-dresden.de/plip-web/plip) (Salentin, 2015) was used to analyze the type of interactions between protein receptors and ssDNA aptamer molecules. The discovery studio visualizer was used for preparing the 3D-interaction surfaces between the receptors and ligands.
To conjugate the DNA aptamer with the MSNs, the disulfide binding which is one of the strongest chemical bindings, was selected. To bind the DNA aptamer to MSNs, the oligo synthesis order was prepared by a 3’-thiol group modification. Previously, it has been indicated that 3’ chemical modification does not affect the structure of these G-quadreplex aptamers against protein A of S. aureus (Stoltenburg, 2016). Synthesis of these modified oligonucleotide sequences was performed by the Generay Biotechnology Company (China), and HPLC was selected as the method of purification.
2.3 Synthesis and characterization of the MSNs
In order to synthesize the MSNs, a method for rapid synthesis of well-ordered MSNs with a narrow pore size distribution introduced by Yun-yu et al. was used (Zhou et al., 2012), with some modifications. In this method, sodium silicate was used as the silica source and cetyltrimethyl ammonium bromide (CTAB) was used as the template for the pore formation of nanoparticles. First, 100 mL of sodium silicate solution (5 wt.%) was prepared and then CTAB was dissolved in 40 mL of 2 M HCl solutions, where the weight ratio of sodium silicate/CTAB was 5 to 1.5. Then, both solutions were added to 100 mL of distilled water under vigorous stirring. In order to adjust the pH value at 3, 1 M HCl was added in a dropwise manner. After stirring for 10 min, the mixture was placed in an oven at 40 oC for 24 h. Then, the precipitate was centrifuged and washed several times until the pH value reached the pH of distilled water, and dried at 80 oC. Finally, the product was calcined at 600 oC in an electric furnace for 3 h under air atmosphere. To characterize the synthesized nanoparticles, X-ray diffraction (XRD), and field-emission scanning electron microscopy (FE-SEM) analyses were performed. The Brunauer-Emmett-Teller (BET) method was used to investigate the available surface area of the synthesized nanoparticles.
2.4 Conjugation of the aptamers with MSN
To conjugate MSNs with the selected anti-protein A aptamers, first, 50 mg of the MSNs was dispersed in 10 mL of distilled water and sonicated at room temperature for 20 min. Then, 70 µL of 3-MPTMS as a linker agent was added to the suspension, and pH was adjusted to 4 and stirred at 25 oC for 3 h to prepare SH-MS nanoparticles.
After the preparation of SH-MS nanoparticles, the G-quadreplex ssDNA aptamers were conjugated to the SH-MS nanoparticles by applying 2 mM CaT in 10 mM MES buffer for 1 min, according to the method used by Zhu et al. (Zhu, 2009).
2.5 Nanoparticle conjugates characterization
To initially inspect the conjugation between the selected aptamer and the MSNs, first, the agarose gel retardation assay was carried out on 1.5 % agarose gel. By this assay, it was aimed to investigate if the DNA content of aptamer conjugated with MSNs shows any change in the movement pattern compared to the free aptamers.
Dynamics light scattering (DLS) was used to investigate the hydrodynamic size of nanoparticle components in the suspension, and Zeta potential was used to study the electrical changes on the surface of the nanoparticles by a Horiba SZ-100 series analyzer.
2.6 Loading and release of Ampicillin in MSN-aptamers conjugate
Initially, 100 mg of the MSNs-Aptamer system and 100 mg of Ampicillin were dissolved in 100 mL of deionized water and stirred at room temperature for 24 h. After that, the suspension was centrifuged at 9000g for 15 min to sediment the nanoparticles containing ampicillin in their pores. The optical density (OD) of the supernatant was investigated to evaluate the drug absorption by nanoparticles. In order to assure the loading of the ampicillin in MSNs, FTIR, DLS, and zeta potential analyses were carried out. After confirming the presence of Ampicillin in MSN-aptamers, the drug release assay was performed using a 12 KDa molecular cut-off dialysis bag in phosphate-buffered saline (PBS) at a pH of 6.5 (similar to that of S. aureus biofilm) for 48 h. Samples were taken every 2 h during the first 12 h, then they were collected every 6 h until 48 h. The OD of the collected samples was compared to the standard calibration curve of Ampicillin to assay the release of the drug by MSNs.
2.7 Biofilm suppression assay
2.7.1 Biofilm inhibition microtiter plate (MTP) assay
Investigating the biofilm inhibition effect of the three-component system on the biofilm of S.aureus (ATCC 6538) was carried out on the 96-wells culture plates. For this purpose, S.aureus was cultured at 37 °C overnight, then cultures were diluted at a ratio of 1:100 in a fresh nutrient broth medium. 200 μL of the fresh culture was inoculated into every three wells of sterile 96-well flat-bottomed polystyrene plate (SPL, South Korea).
To suppress the S.aureus biofilm formation, different doses of MSN, MSN-Aptamer, and MSN-Aptamer-Ampicillin (10, 25, 50, 75, and 100 µg/mL) were added to each well. Control wells contained fresh nutrient broth media. The plates were placed into a shaker incubator for 48 h at 37 °C.
After 48 h, the media was discharged and each well was washed three times with 300 μL of PBS. The wells were dried at temperature. The attached biofilm was fixed using 300 mL of 99 % methanol, and after 15 min, the plates were emptied and left to be dried. The plates were stained with 250 μL of 1 % crystal violet for 10 min. The stain was poured out and the wells were washed with distilled water to remove the extra dye. Finally, 200 μL of acetic acid (33 %) was added to each well and the OD value of each well was inspected at 630 nm, using a microplate reader (Elx808, BioTek, USA).
2.7.2 Anti-Biofilm activity of nanosystem on mice bone
S. aureus biofilm formation on mice bone
To investigate the degradative effects of the composed nanosystem to suppress the biofilm formation of S. aureus on mice bone, initially, the flat bones of mice (skull) were harvested by sacrificing six weeks old male Balb/c mice and were cleaned from extra tissues using a sharp scalpel. The bones were cut at the 5 mm × 5 mm sizes and were autoclaved for 15 min at 121 oC to completely sterile them from any bacteria. Sterile bones were stored at 4 oC before performing the biofilm suppression assay.
In order to form the biofilm on the surface of mice bones, first, an overnight culture was prepared by culturing S. aureus in a nutrient broth medium. Afterward, bones were placed in a 24-wells culture plate containing 400 µL of fresh media and 100 µL of overnight culture. After 24 h, the bones were moved to a new well containing 500 µL of fresh medium to only cultivate the ones attached to the surface of the bone. The process was performed for 72 h, until the clear biofilm on the sample was witnessed on the bone surface by light microscopy.
Bone biofilm degradation by composed nanosystem
To suppress the S.aureus biofilm on the surface of bone tissue, different doses of MSN, MSN-Aptamer, and MSN-Aptamer-Ampicillin (10, 25, 50, 75, and 100 µg/mL) were added to bones with 72 h biofilm and placed in 37 oC shaker incubator for 48 h. Then, the bones were washed with fresh media and placed into a new plate, so that only the microbes on the bone biofilm be presented in the plate. In the next step, the bones were washed with PBS (pH 7.2) three times, then they were dried at room temperature. Afterward, 300 µL of the 1 % crystal violet for 10 min, and were washed with deionized water three times. In the next step, the bones were exposed to 300 µL of 98 % ethanol to remove all the biofilm from the bone surface. The bones were removed from the plate and the results were investigated using an ELISA microplate reader at 630 nm. The results were provided by comparing the mean OD of the test groups with that of the control group.
Light microscopy
To study the S. aureus presence by light microscopy, the bones were dyed using Gram methods and investigated with a ×100 lens. The presence of biofilm was expected to be seen as some dark blue microbial populations on the bone samples.
SEM analysis
Bone samples with 72 h biofilm and treated ones with 100 µg/mL of the nanosystem were subjected to SEM investigation. In order to prepare the samples for SEM analysis, first, the bones (containing biofilm and treated with nanosystem) were placed into a 2.5 % glutaraldehyde solution for 4 h, then the samples were passed through a graded solution of ethanol (10, 20, 30, 40, 50, 60, 70, 80, 90, and 98 %) for 10 min, then they were dried at room temperature overnight, before SEM studies.
2.8 Cell viability assay
To study the potential toxicity of the nanosystem for human cells, the cell viability assay was carried out using the MTT 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide reagent on MCF7 cell line. To do this, first, MCF7 cells were cultivated in 25 T flasks, containing DMEM medium, fetal bovine serum (FBS), and pen-strep antibiotic. 20,000 cells were seeded in each well of the 96-wells plate. Afterward, the cells were exposed to the composed nanosystem for 48 h at different concentrations (i.e. 10, 25, 50, 75, and 100 µg/mL). The result of toxicity was acquired by comparing the count of treated cells with the non-treat group.
2.9 Statistical assay
GraphPad Prism 8.02 program was used for statistical analysis of data. All of the values are presented as mean ± standard deviation (SD). Two-way analysis of variance (ANOVA) was used for statistical assays. Values with P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****) were considered statistically important. Each experiment was at list repeated three times.
3 Results
3.1 Selection of the specific aptamer against S. aureus
Mfold analysis of the truncated structures indicated that the G-quadreplex construct does not change until 53 nucleotides truncate form. Accordingly, molecular docking was carried out on aptamer sequences from PA76 to PA53, which is the last G-quadreplex DNA aptamer. The results of molecular docking between the proposed aptamer structures and domain C of protein A are provided in Table 1. For more information regarding model and cluster analysis of in silico assay, please refer to Figs. S1 and S2, respectively.
| Aptamer | HADDOCK score | Cluster size | RMSD from the overall lowest structure | Vander Waals energy (kcal/mole) | Electrostatics energy (kcal/mole) | Desolvation energy (kcal/mole) | Restraint violation energy (kcal/mole) | Buried surface area (Ao2) | Z-score |
|---|---|---|---|---|---|---|---|---|---|
| PA76 | −1.8 | 33 | 11.9 | −88.8 | −286.1 | 6.1 | 1381.9 | 2422.9 | −2.1 |
| PA75 | 5.9 | 4 | 11.4 | −80.4 | −436.2 | 14.9 | 1586.6 | 2394.7 | −1.1 |
| PA74 | −20 | 17 | 15.6 | −88.1 | −352.9 | 6.3 | 1323.5 | 2667.6 | −1.8 |
| PA73 | −3.3 | 14 | 15.8 | −77.9 | −323.4 | 5.1 | 1341.9 | 2447.4 | −1.8 |
| PA72 | −13.7 | 9 | 6 | −70.7 | −343.1 | 8.2 | 1175.5 | 2322.5 | −1.7 |
| PA71 | −31.6 | 19 | 1.8 | −79.6 | −418.6 | 18.6 | 1131 | 2597.8 | −2.4 |
| PA70 | −32 | 15 | 11.6 | −100.5 | −272.6 | 12.7 | 1102.7 | 2807.6 | −2.3 |
| PA69 | −8 | 6 | 11.9 | −67.7 | −330.5 | 20.3 | 1054.9 | 2187.3 | −1.5 |
| PA68 | −28.2 | 9 | 6.1 | −70.9 | −517.3 | 32.3 | 1138.7 | 2547 | −1.5 |
| PA67 | −32.5 | 5 | 12.9 | 71.7− | −448.3 | 27.1 | 1018.0 | 2367.2 | −1.6 |
| PA66 | −86.4 | 26 | 2.1 | −91.7 | −556.5 | 33.3 | 833.5 | 3221.1 | −1.8 |
| PA65 | −60.7 | 10 | 17.9 | −92.8 | −377.4 | 32.4 | 751 | 2741.9 | −1.9 |
| PA64 | −43.7 | 18 | 13.3 | −101 | −285.6 | 17.7 | 968.3 | 2851.7 | −1.4 |
| PA63 | −98.1 | 21 | 1.0 | −102 | −573.2 | 38.4 | 783.4 | 3056.1 | −2.3 |
| PA62 | −15.3 | 44 | 18 | −82.9 | −250.6 | 5.8 | 1119.8 | 2200.4 | −1.4 |
| PA61 | −70 | 17 | 12.5 | −98.1 | −448.4 | 30.6 | 827.5 | 2753.4 | −1.9 |
| PA60 | −35 | 34 | 12.8 | −83.2 | −324.6 | 9.2 | 1033.1 | 2192.8 | −1.5 |
| PA59 | −18.5 | 13 | 14.6 | −73.4 | −319.4 | 21.1 | 977.6 | 2287.4 | −1.4 |
| PA58 | −52.2 | 19 | 13.3 | −101.6 | −225.7 | 12.5 | 820.3 | 2683.9 | −1.9 |
| PA57 | −72.2 | 5 | 1.4 | −96.5 | −387.1 | 25.9 | 758 | 2783.3 | −2.1 |
| PA56 | −23.5 | 15 | 14.2 | −77.9 | −303.7 | 20.3 | 98.1 | 2273.9 | −1.8 |
| PA55 | −97.5 | 18 | 1.1 | −87.6 | −549.8 | 39.9 | 601.4 | 3011.6 | −2.2 |
| PA54 | −77.3 | 5 | 1.3 | −101.8 | −314 | 26 | 616.5 | 2911.3 | −1.9 |
| PA53 | −96.4 | 21 | 1.2 | −98.9 | −449.1 | 38.1 | 543 | 3253.4 | −2 |
Results of molecular docking indicated that PA63 has the highest docking score, and the highest Vander Waals energy of −102 kcal/mole, which makes it a suitable candidate for being a bio-recognition element against S. aureus biofilm. This construct has the sequence of 3’-ATACCAGCTTATTCAATTAGCAACATGAGGGGGATAGAGGGGGTGGGTTCTCTCGGCTACAAT-5’ with a molecular weight of 19,898.60 g/mol and 47 % GC content. Mfold predicted a ΔG = −2.25 kcal/mol. Fig. 2 indicates the secondary and 3D structure of this aptamer.
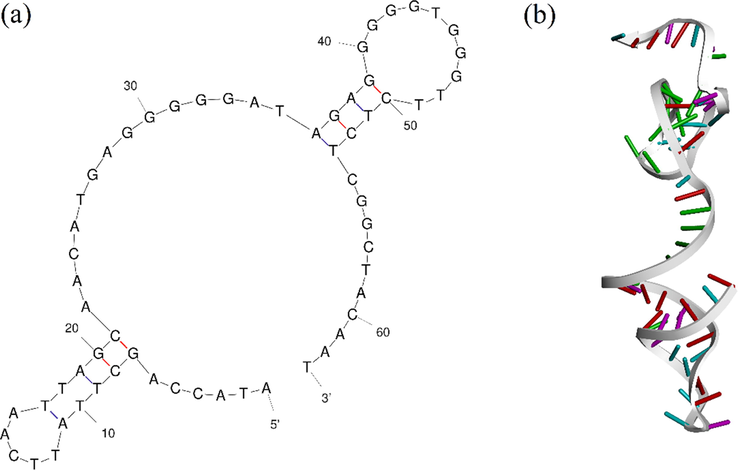
- PA63 G-quadreplex aptamer (a) Secondary structure of PA63 oligonucleotide construct, and (b) Tertiary structure of PA63 oligonucleotide sequence.
Fig. 3 represents the protein A-PA63 complex regarding different surfaces of interaction. The detail of interactions by PLIP webserver is provided in Table S1. Although the results of PLIP indicated that there is no direct interaction with residues after 58 nucleotides, the highest docking score was for PA63.In this case, the reason could be that the addition of the 5 next nucleotides could affect the 3D structure and overall energy. Accordingly, PA63 was selected as the optimum construct for targeting protein A.

- Protein A-PA63 Aptamer complexes surface of interactions with regards to (a) Aromatic interactions, (b) Intercalated charges, (c) Hydrogen bonds, (d) Hydrophobicity, (e) Ionizablity, and (f) Solvent accessibility.
3.2 Synthesis and characterization of MSNs
Fig. 4a shows the SEM micrograph of the synthesized MSNs. In addition, the results of the N2 adsorption/desorption test (Figure 4B) indicated that the synthesized MSNs had a specific surface area (SBET) of 850.6 m2/g, total pore volume of 1.3875 cm3/g, and mean pore diameter of 6.5 nm. Also, the presence of a peak at about 2o in the low-angle XRD pattern (Fig. 4c) confirms the mesoporous characteristic of the synthesized particles with an amorphous structure. Fig. 4d presents the result of normal angle XRD analysis of particles.
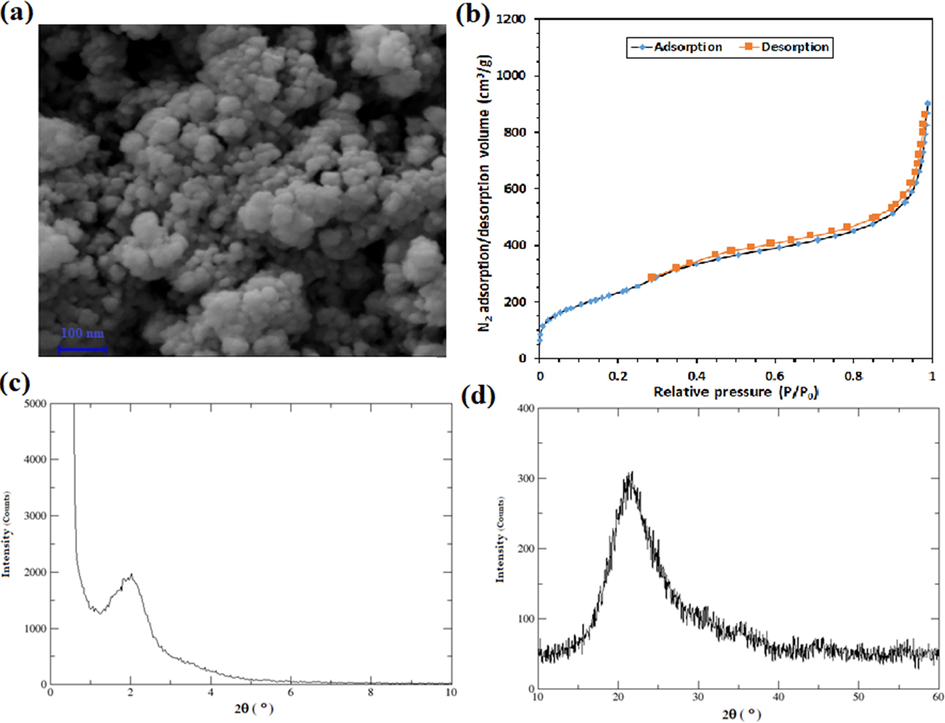
- The results of the characterizations of the synthesized MSNs by (a) FE-SEM analysis, (b) N2 adsorption/desorption assay, (c) low-angle XRD test, and (d) normal angle XRD test.
3.3 Conjugation of the aptamer with MSN
To confirm the formation of S = S bind between the selected aptamer with MSNs, first, the gel retardation assay was used through agarose gel electrophoresis. This assay indicated that despite the free aptamers, MSNs-APT conjugates move slower in the agarose gel, indicating that the MSNs conjugation with aptamers was successful (Fig. S3).
DLS test was used to investigate the changes in the hydrodynamic diameter of nanoparticles. The results of this investigation showed that the mean hydrodynamic diameter of nanoparticles was 248.8 ± 48.7 for MSN nanoparticles. After the thiolation of MSNs, the diameter was changed to 286.0 ± 15.8 nm, meaning that the particles were more aggregated. Upon conjugation with PA63 aptamers, the hydrodynamic diameter was changed to 185.7 ± 8.6 which means that the addition of aptamers reduced the aggregation of nanoparticles by causing a barrier between those particles.
Regarding the Zeta potential assay, it was shown that the Zeta potential of MSNs was −53.8 mV. These values were −54.3 mV for SH-MSNs, respectively. This indicates that the addition of the thiol group slightly reduced the zeta potential of MSNs. After conjugation of the aptamer to the structure, the zeta potential value was reduced to −40.9 mV. (For more information regarding DLS and Z-potential analyses, please refer to Fig. S4)
3.4 Drug loading and release assay
To investigate the loading of Ampicillin into pores of MSNs, initially, the OD of supernatant from the loading solution was measured after sedimentation of the nanoparticles by centrifuging at 7000g for 10 min. The result indicated that 20 mg of the drug was loaded into the pores of nanoparticles.
The results of the DLS test indicated that after loading of Ampicillin, the size of particles was increased to 326.7 ± 15.9 nm (Fig. S4a), showing that some of the drug molecules are present at the surface of nanoparticles. The zeta potential value was also changed to −72.9 mV (Fig. S4b).
FTIR analysis was carried out on MSNs, and the three-components system. These results showed that after the loading of Ampicillin, sensible changes could be witnessed on the FTIR spectrum (Fig. 5a). In order to analyze the release of Ampicillin from the MSN nanoparticles, the dialysis assay was carried out. This analysis indicated that almost 30 % of the absorbed drug was released into the medium upon 48 h (Fig. 5b).
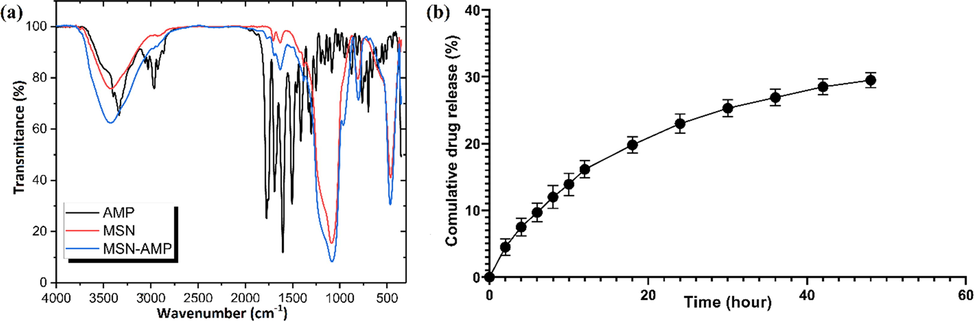
- Loading and release assays of Ampicillin into the nanosystem, (a) The results of FTIR-DR analysis of MSNs, Ampicillin, and the ampicillin-loaded nanosystem, and (b) Cumulative Ampicillin release from the three-components nanosystem for 48 h.
3.5 Biofilm inhibition assay
3.5.1 MTP assay
The biofilm prevention assay was performed by microtiter plate assay using a 96-wells plate. The results (Fig. 6a) showed that MSNs alone do not pose any significant anti-biofilm activity against the formation of biofilm (less than 10 % in all concentrations), but MSNs-APT, MSNs-AMP, and MSNs-APT-AMP pose a significant anti-biofilm formation activity. The three-component nanosystem showed the highest activity in all concentrations compared to all the other groups (P < 0.0001), and this effect was increased dose-dependently, in a way that in 100 µg/mL concentration, the biofilm was mostly suppressed in 72 h (more than 90 %).
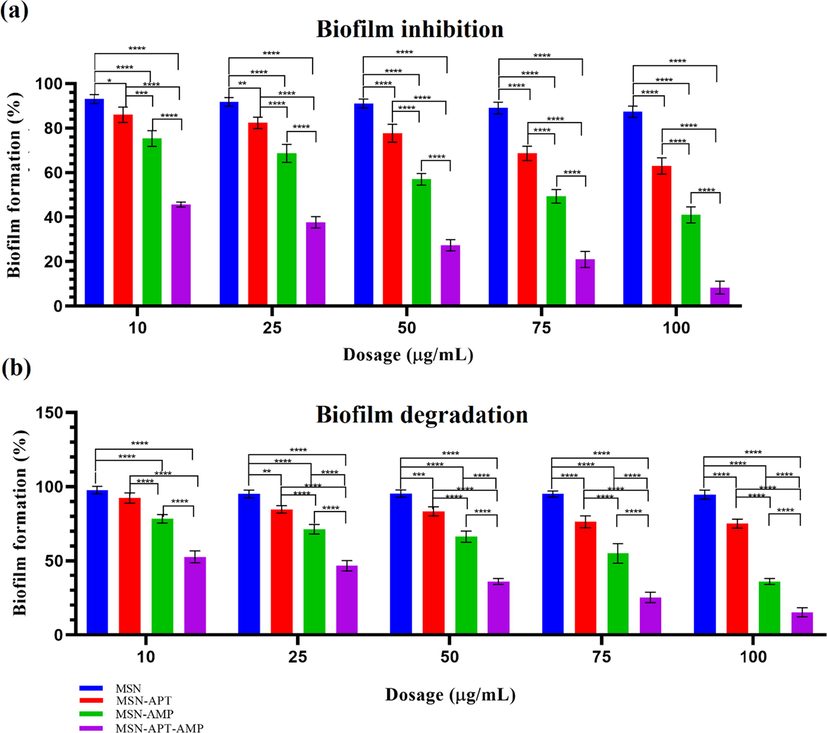
-
(a) Preventing biofilm formation through microtiter plate assay using 10, 25, 50, 75, and 100 µg/mL concentrations of MSNs, MSNs-APT, MSNs-AMP, and three-component system. (b) Suppression of the formed biofilm on the surface of mice skull bone using 10, 25, 50, 75, and 100 µg/mL concentrations of MSNs, MSNs-APT, MSNs-AMP, and MSNs-APT-AMP. Values with P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****) were considered statistically important. Each experiment was at list repeated three times.
3.5.2 Bone biofilm degradation
MTP assay of degrading formed biofilm on mice bones
The results of investigating MSNs, MSNs-APT, MSNs-AMP, and the three-component system on suppressing the formed 72 h biofilm of the mice bone indicated that MSNs do not provide any significant effect with any of its selected dosages (10, 25, 50, 75, and 100 µg/mL) on the biofilm of S. aureus and the bacterial growth was continued (Fig. 6b). In the case of MSNs-APT, only 10 µg/mL did not have any significant effect compared to the MSNs group, while 25 (P < 0.01), 50 (P < 0.001), 75, and 100 µg/mL (both P < 0.0001) could significantly suppress the biofilm. MSNs-AMP provided a higher anti-biofilm effect than MSNs and MSNs-Aptamer in all dosages (P < 0.0001), and the three-component system showed the highest efficacy, which was significant compared to all of the other groups (P < 0.0001).
Light microscopy
Results of light microscopy using Gram staining indicated the presence of S. aureus biofilm on the surface of mice bone in 72 h non-treated group (Fig. S5a), while there was no significant biofilm on the surface of the bone after 48 h treatment with 100 µg/mL of the three-component system (Fig. S5b).
SEM microscopy
The results of the SEM study (Fig. 7) indicated the formation of a strong biofilm on the surface of mice bone in 72 h (part a), while a weak (part b) and almost no biofilm (part C) were seen on the surface of the treated group with 100 µg/mL of the three-component system after 48 h. Fig. 7, part D shows the presence of the three-component nanosystem on the mice's skull surface.
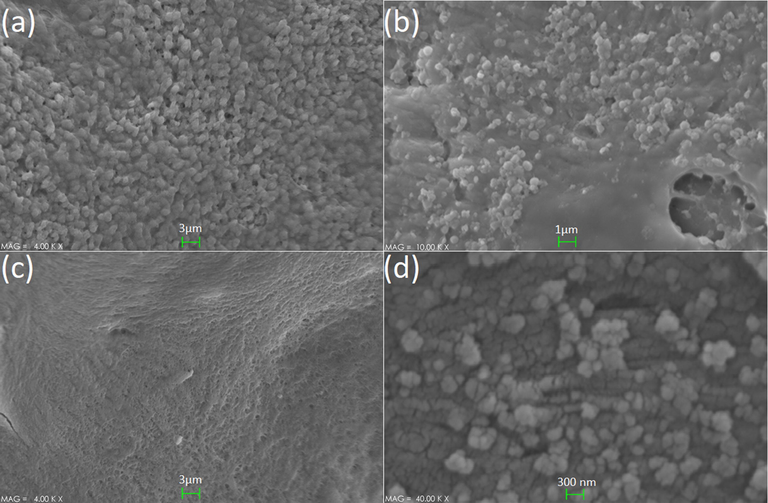
- The results of SEM investigations of the samples, (a) strong biofilm (none treated), (b) moderate biofilm (24 h treated), (c) weak/almost none biofilm (48 h treated), and (d) the presence of three-component nanosystem on the mice bone surface.
3.6 Cell viability assay
The cell viability assay by using the MTT method indicated that neither of MSNs, MSNs-AMP, MSNs-APT, and the three-component pose any significant (P < 0.05 was considered statistically meaningful in two-way ANOVA) toxicity in the 10, 25, 50, 75, and even 100 µg/mL doses for the MCF-7 cells. Fig. 8 shows the results of the cell toxicity assay of the mentioned components on MCF7 cells.
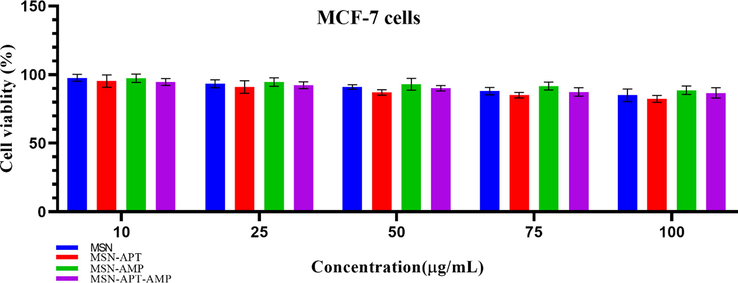
- Results of 48 h MTT assay on MCF-7 cell line with 10, 25, 50, 75, and 100 µg/mL concentrations of MSNs, MSNs-Aptamer, and three-component system. No significant toxicity (p-value < 0.05) was witnessed in any of the studied groups. Each experiment was at list repeated three times.
4 Discussion
Bone biofilm and osteomyelitis are regarded as deadly infections that mostly happen after open bone fractures and/or surgical interventions. Such infections are mostly caused by opportunist pathogens, among which S. aureus is regarded as a leading agent. The emergence of multi-drug resistance and mutual infection with the other species has made the treatment of this agent even more complicated. So far, many attempts have been given toward developing an effective means to suppress this clinical pathogen, among them novel methods such as the application of nanotechnology methods are highlighted for providing astonishing outcomes in the laboratory and bedside (Gao, 2021; Machado, 2010).
During the last decade, the emergence of sciences such as novel drug delivery methods have completely revolutionized the scientific insight toward developing therapeutics against a variety of disorders, including cancer therapy infections with bacterial or viral agents and even tumor metastasis (Laffleur and Keckeis, 2020; Mousazadeh, 2022). The application of MSNs has been considered by different drug delivery studies, due to their high specific surface area (Stephen, 2021). Many studies have shown that these particles could be considered suitable drug carriers, along with the potential to specifically define their targets by surface decorating of these particles with bio-recognition elements (Barui and Cauda, 2020).
Previously, the use of three-component systems of MSN-Aptamer-drug has been used against various disorders. For example, a study by Li et al. (2012) reported the application of polyvalent MSNs-Aptamers to target breast cancer cells. They provided their aptamer through SELEX assay and synthesized their MSNs using TEOS and CTAB. They used TSPMP to prepare MSNs-pho and functionalized their aptamers by cross-linker Sulfo-SMCC. Their study indicated that the efficacy of doxorubicin delivery to the breast cancer cells was considerably increased using such a three-component system.
Another study by Xie et al. (2016) investigated the efficacy of EpCAM aptamer-coated MSNs to target colon cancer cells. In their study, they used these particles to deliver doxorubicin to cancer cells. Their results suggested that the application of such a three-component system could significantly increase the therapeutic index while reducing the side effects of drugs by restricting exposure toward high drug doses. In case of the current study, the used three-component system does also reduce exposure toward high doses of antibiotics, which are regularly used to battle osteomyelitis (Rao et al., 2011). It is a well-known fact that the application of high doses of antibiotics could cause various problems; such as developing multidrug-resistant strains like MRSA or harming the normal microbiota (Baptista, 2018). Therefore, developing any means of drug delivery system that restricts the high load uses of antibiotics is helpful to medical society and care-seekers.
Previously, application of the three-component system of MSNs-Gentamycin (antibiotic) and a bacterial targeting peptide UBI29-41has been performed to battle the biofilm of S. aureus. Yang et al. reported the high efficacy of this system for suppressing intracellular S. aureus and associated infection through in vitro and in vivo analysis (Yang, 2018). In our study, DNA aptamers were used to target S. aureus, instead of a peptide sequence. The advantages of using aptamers rather than peptides and antibodies have been discussed in the literature (Li, 2021). In practice, the expenses of peptide synthesis and purification are higher than DNA aptamer sequences. Moreover, the application of DNA aptamers is causing fewer side effects such as allergenicity and toxicity compared to peptides. Accordingly, using nucleic acid aptamers rather than peptides could provide more satisfying results with a high potential for future medical applications.
One of the advantages of the three-component system in this study was using a G-quadreplex aptamer, instead of the regular aptamer constructs. Since the biofilm environment of S. aureus is slightly acidic (Fernández, 2021), it does not seem practical to use regular aptamers that are sensitive toward pH changes. Accordingly, the application of a G-quadruplex aptamer which is more tolerant toward rough conditions; such as high temperature and pH changes (Platella, 2017), could increase the efficacy of the drug delivery to the site of infection, compared with using regular ssDNA aptamers.
In the current study, we investigated the efficacy of a novel three-component system, composed of a G-quadreplex aptamer as the bio-recognition element, MSNs as the drug carrier, and Ampicillin as the loading drug inside of the particles. This system aimed to reduce the unnecessary exposure of the host cells toward high dosages of antibiotics that is required for the suppression of osteomyelitis. In this study, we used bioinformatics methods to investigate the molecular interactions between the previously developed aptamer for binding protein A of S. aureus and we spotted the sequence and length of the aptamer with the highest binding affinity toward its proposed molecular target.
The results of the current study confirmed that this three-component system could considerably increase the efficacy of drug delivery to the biofilm of S. aureus, without posing any significant toxicity for eukaryotic cells. It was indicated that this system provides higher efficacy in preventing the formation of S. aureus biofilm, compared to the destruction of formed biofilm on the surface of the bone. It was indicated that the activity of this system increases by elevating its dosage to 100 µg/mL, and this concentration did not show any significant toxicity for MCF-7 cells in 48 h.
Application of the G-quadreplex aptamer rather than the simple ones was one of the advantages of this system because it could tolerate the acidic pH of S. aureus biofilm that could destroy the regular ssDNA aptamers. Moreover, it was shown that this aptamer, even in absence of a loaded drug in MSNs, could suppress the biofilm formation of this bacterium, which could be due to structural inhibition of protein A, that is a necessary component for biofilm formation.
Finally, it needs to be mentioned that although this nanosystem showed efficacy in the suppression of S. aureus biofilm in vitro, far more studies including in vivo investigations are required to be carried out to confirm its practical application for any potential use in humans. Moreover, it is necessary to perform more investigations by modifying the components such as the carrier and/or cargo drug to compare the results and confirm what system may present more practical merits to be used for future studies or possibly human application in hospitals.
5 Conclusion
The results provided by this study indicated that the proposed nanosystem of MSN-Aptamer-Ampicillin could provide an effective platform for the gradual release of antibiotics, which could suppress the S. aureus biofilm in bone tissue. The importance of such a system is in the prevention of osteomyelitis and could be used in the site of open bone fractures or after long-lasting surgeries such as hip or knee bone surgeries as surgical pads. Further in vivo studies are required prior to suggesting this nanosystem for clinical applications.
Compliance with ethical standards
Due to the involvement of mice sacrifice in the study, a moral code was provided as IR.UI.REC.1398.061by the ethical committee of scientific researches, University of Isfahan, Isfahan, Iran who supervised the moral aspect of this study.
CRediT authorship contribution statement
Mohammad Moradi: Conceptualization, Data curation, Formal analysis, Investigation, Software, Writing - original draft. Hassan Mohabatkar: Supervision. Mandana Behbahani: Supervision. Ghasem Dini: Formal analysis, Writing – review & editing, Supervision.
Acknowledgment
This article is the output of a Ph.D. thesis by Mohammad Moradi, which was approved by the University of Isfahan, and received financial support from this University. We wish to thank them for their kind supports.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. There is no conflict of interests.
References
- Nano-strategies to fight multidrug resistant bacteria—“A Battle of the Titans”. Front. Microbiol.. 2018;9:1441.
- [Google Scholar]
- Nanotechnology-Based Targeted Drug Delivery: An Emerging Tool to Overcome Tuberculosis. Adv. Therap.. 2021;4(1):2000113.
- [Google Scholar]
- A Pseudomonas aeruginosa Antimicrobial Affects the Biogeography but Not Fitness of Staphylococcus aureus during Coculture. Mbio. 2021;12(2):e00047-e121.
- [Google Scholar]
- Multimodal decorations of mesoporous silica nanoparticles for improved cancer therapy. Pharmaceutics. 2020;12(6):527.
- [Google Scholar]
- Using Chou’s general pseudo amino acid composition to classify laccases from bacterial and fungal sources via Chou’s five-step rule. Appl. Biochem. Biotechnol.. 2020;190(3):1035-1048.
- [Google Scholar]
- In silico design of quadruplex aptamers against the spike protein of SARS-CoV-2. Elsevier; 2021.
- A multicenter randomized placebo controlled trial of rifampin to reduce pedal amputations for osteomyelitis in veterans with diabetes (VA INTREPID) BMC Infect. Dis.. 2020;20(1):1-12.
- [Google Scholar]
- The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc.. 2010;5(5):883-897.
- [Google Scholar]
- Environmental pH is a key modulator of Staphylococcus aureus biofilm development under predation by the virulent phage phiIPLA-RODI. ISME J.. 2021;15(1):245-259.
- [Google Scholar]
- Combination of kaempferol and azithromycin attenuates Staphylococcus aureus-induced osteomyelitis via anti-biofilm effects and by inhibiting the phosphorylation of ERK1/2 and SAPK. Pathogens Disease. 2021;79(8) p. ftab048
- [Google Scholar]
- Potentials of nanotechnology in treatment of methicillin-resistant Staphylococcus aureus. Eur. J. Med. Chem.. 2021;213:113056
- [Google Scholar]
- NANOG Decoy Oligodeoxynucleotide-Encapsulated Niosomes Nanocarriers: A Promising Approach to Suppress the Metastatic Properties of U87 Human Glioblastoma Multiforme Cells. ACS Chem. Neurosci.. 2020;11(24):4499-4515.
- [Google Scholar]
- Hybrid of niosomes and bio-synthesized selenium nanoparticles as a novel approach in drug delivery for cancer treatment. Mol. Biol. Rep.. 2020;47(9):6517-6529.
- [Google Scholar]
- Immuno-informatics analysis and expression of a novel multi-domain antigen as a vaccine candidate against glioblastoma. Int. Immunopharmacol.. 2021;91:107265
- [Google Scholar]
- Formulation and biocompatibility of microemulsion-based PMBN as an efficient system for paclitaxel delivery. J. Appl. Biotechnol. Rep.. 2021;8(1)
- [Google Scholar]
- González-García, S. et al., 2021. Factors of Nasopharynx that Favor the Colonization and Persistence of Staphylococcus aureus. In: Pharynx-Diagnosis and Treatment. IntechOpen.
- Concepts in wound irrigation of open fractures:‘Where we came from, and where are we now? J. Clin. Orthopaedics Trauma 2021101638
- [Google Scholar]
- In silico study of the structure and ligand interactions of alcohol dehydrogenase from Cyanobacterium Synechocystis sp. PCC 6803 as a key enzyme for biofuel production. Appl. Biochem. Biotechnol.. 2020;192(4):1346-1367.
- [Google Scholar]
- Computational approach to analyze isolated ssDNA aptamers against angiotensin II. J. Biotechnol.. 2016;230:34-39.
- [Google Scholar]
- High affinity truncated aptamers for ultra-sensitive colorimetric detection of bisphenol A with label-free aptasensor. Food Chem.. 2020;317:126459
- [Google Scholar]
- Evaluation of STAT3 decoy oligodeoxynucleotides' synergistic effects on radiation and/or chemotherapy in metastatic breast cancer cell line. Cell Biol. Int.. 2020;44(12):2499-2511.
- [Google Scholar]
- Bone Regeneration in rat using a gelatin/bioactive glass nanocomposite scaffold along with endothelial cells (HUVECs) Int J Appl Ceram Technol. 2018;15:1427-1438.
- [CrossRef] [Google Scholar]
- Advances in drug delivery systems: Work in progress still needed? Int. J. Pharmac.: X. 2020;2
- [Google Scholar]
- Polyvalent mesoporous silica nanoparticle-aptamer bioconjugates target breast cancer cells. Adv. Healthcare Mater.. 2012;1(5):567-572.
- [Google Scholar]
- Nucleic acid aptamers for molecular diagnostics and therapeutics: advances and perspectives. Angew. Chem. Int. Ed.. 2021;60(5):2221-2231.
- [Google Scholar]
- A novel aptamer-based histochemistry assay for specific diagnosis of clinical breast cancer tissues. Chin. Chem. Lett.. 2021;32(5):1726-1730.
- [Google Scholar]
- Lyu, C., Khan, I.M., Wang, Z., 2021. Capture-SELEX for aptamer selection: A short review. Talanta, pp. 122274.
- Targeted drug delivery strategies for precision medicines. Nat. Rev. Mater.. 2021;6(4):351-370.
- [Google Scholar]
- Evolving concepts in bone infection: redefining “biofilm”, “acute vs. chronic osteomyelitis”, “the immune proteome” and “local antibiotic therapy”. Bone Res.. 2019;7(1):1-18.
- [Google Scholar]
- A concise IN silico prediction report OF a potential PRION-like domain IN SARS-COV-2 polyprotein. J. Microbiol. Biotechnol. Food Sci. 2021:e4813-e.
- [Google Scholar]
- Anticancer evaluation of methotrexate and curcumin-coencapsulated niosomes against colorectal cancer cell lines. Nanomedicine. 2022;17(4):201-217.
- [Google Scholar]
- A comprehensive review of bacterial osteomyelitis with emphasis on Staphylococcus aureus. Microb. Pathog. 2020104431
- [Google Scholar]
- Nucleic acid aptamers: clinical applications and promising new horizons. Curr. Med. Chem.. 2011;18(27):4206-4214.
- [Google Scholar]
- Complete ablation of tumors using synchronous chemoradiation with bimetallic theranostic nanoparticles. Bioact. Mater.. 2022;7:74-84.
- [Google Scholar]
- Aptamer-gated nanoparticles for smart drug delivery. Pharmaceuticals. 2011;4(8):1137-1157.
- [Google Scholar]
- G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochimica et Biophysica Acta (BBA)-Gen. Subj.. 2017;1861(5):1429-1447.
- [Google Scholar]
- Thermodynamic and first-principles biomolecular simulations applied to synthetic biology: Promoter and aptamer designs. Mol. Syst. Des. Eng.. 2018;3(1):19-37.
- [Google Scholar]
- Staphylococcal trafficking and infection—from ‘nose to gut’and back. FEMS Microbiol. Rev. 2021
- [Google Scholar]
- Treating osteomyelitis: antibiotics and surgery. Plast. Reconstr. Surg.. 2011;127:177S-187S.
- [Google Scholar]
- PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res.. 2015;43(W1):W443-W447.
- [Google Scholar]
- Exploring the role of mesoporous silica nanoparticle in the development of novel drug delivery systems. Drug Deliv. Trans. Res. 2021:1-19.
- [Google Scholar]
- G-quadruplex aptamer targeting Protein A and its capability to detect Staphylococcus aureus demonstrated by ELONA. Sci. Rep.. 2016;6(1):1-12.
- [Google Scholar]
- In vitro selection and interaction studies of a DNA aptamer targeting protein A. PLoS ONE. 2015;10(7):e0134403.
- [Google Scholar]
- Mid-term clinical results of chronic cavitary long bone osteomyelitis treatment using S53P4 bioactive glass: a multi-center study. J. Bone Joint Infect.. 2021;6(9):413-421.
- [Google Scholar]
- In silico post-SELEX screening and experimental characterizations for acquisition of high affinity DNA aptamers against carcinoembryonic antigen. RSC Adv.. 2019;9(11):6328-6334.
- [Google Scholar]
- Risk factors for surgical site infection following spine surgery: efficacy of intraoperative saline irrigation. J. Neurosurg.: Spine. 2010;12(5):540-546.
- [Google Scholar]
- Predicting the causative pathogen among children with osteomyelitis using Bayesian networks–improving antibiotic selection in clinical practice. Artif. Intell. Med.. 2020;107:101895
- [Google Scholar]
- EpCAM aptamer-functionalized mesoporous silica nanoparticles for efficient colon cancer cell-targeted drug delivery. Eur. J. Pharm. Sci.. 2016;83:28-35.
- [Google Scholar]
- Bacteria-targeting nanoparticles with microenvironment-responsive antibiotic release to eliminate intracellular Staphylococcus aureus and associated infection. ACS Appl. Mater. Interfaces. 2018;10(17):14299-14311.
- [Google Scholar]
- Computational analysis and optimization of carcinoembryonic antigen aptamers and experimental evaluation. J. Biotechnol.. 2019;306:1-8.
- [Google Scholar]
- The effects of Peptide Mel4-coated titanium plates on infection rabbits after internal fixation of open fractures. Arch. Orthop. Trauma Surg. 2021:1-6.
- [Google Scholar]
- Rapid synthesis of well-ordered mesoporous silica from sodium silicate. Powder Technol.. 2012;226:239-245.
- [Google Scholar]
- An efficient cell-targeting and intracellular controlled-release drug delivery system based on MSN-PEM-aptamer conjugates. J. Mater. Chem.. 2009;19(41):7765-7770.
- [Google Scholar]
- Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res.. 2003;31(13):3406-3415.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.104274.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







