Translate this page into:
Bacterial synthesis of magnetic Fe3O4 nanoparticles: Decolorization Acid Red 88 using FeNPs/Ca-Alg beads
⁎Corresponding author at: University of Kashan, Ootb-e Ravandi Blvd, Kashan, Isfahan, Iran. jookar@kashanu.ac.ir (Fereshteh Jookar Kashi)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
A new route to biosynthesis of FeNPs using a novel hyperthermophile bacterial strain isolated from Choghart iron mine. FeNPs/Ca-Alg beads had a high ability to decolorize Acid Red 88 dye. Metabolites decolorization of Acid Red 88 dye by FeNPs/Ca-Alg beads was evaluated using GC/MS analysis. FeNPs indicated great anti-cancer activity. FeNPs/Ca-Alg beads effectively kill pathogenic bacteria.
Abstract
The research studied the bacterial synthesis of magnetic iron oxide nanoparticles (FeNPs), their biological activity, and the biodegradation efficiency of Acid Red 88. An iron-resistant bacterial strain S2 was isolated from Choghart iron mine soil and selected to synthesize FeNPs. The phylogenetic analysis depended on 16S rDNA sequence comparison resolution and showed that strain S2 was 99.68% similar to Bacillus zhangzhouensis. FeNPs were identified by UV–Vis, FTIR, and XRD spectroscopy. SEM-EDX conclusions corroborated the size of FeNPs in the range of 20–70 nm in the rhombus form. The finding indicated that FeNPs had antimicrobial activity on tested bacteria. According to the brine shrimp lethality (BSL) test, FeNPs showed great anticancer activity with LC50 value of 0.5 μg/ml. The biodegradation of Acid Red 88 was investigated separately by FeNPs/Ca-Alg beads and FeNPs powder. FeNPs/Ca-Alg beads decolorized Acid Red 88 with 100% efficiency after 24 h, while FeNPs powder decolorized Acid Red 88 with 100% efficiency after 72 h. Moreover, Ca-Alg beads were applied as control and could not decolorize the dye. Decolorization of Acid Red 88 using FeNPs/Ca-Alg beads was investigated by GC/MS analysis. In this analysis, no mass was found related to the initial mass of Acid Red 88. According to these conclusions, it can be inferred that the dye is completely decomposed. The toxicity of Acid Red 88 dye, and the decolorization products, was assessed using BSL and phytotoxicity test. The conclusions determined that Acid Red 88 dye had toxicity effect. Further, the decolorization products could not kill half of the shrimp due to their low toxicity and did not inhibit the germination of radish seeds.
Keywords
FeNPs
Decolorization
Immobilization
Bacteria
Anticancer
Data availability statement
The datasets generated during and/or analyzed during this research are available from the corresponding author on reasonable request.
1 Introduction
Dyes are combinations often utilized in the textile industry (Akar et al., 2009). Extensive production of these dyes leads to environmental pollution. Therefore, these dyes must be removed from the environment. Among dye removal methods from the textile industry, we can mention chemical, physical, and biological processes. Chemical and physical procedures have high limitations for dye removal owing to their great cost and low performance. However, biological methods are effective for removing dye from colored effluents due to their cost-effectiveness and environmental friendliness (Rajeshwari et al., 2011; Rajendra et al., 2012; Marimuthu et al., 2013). Biological treatment of wastewater containing textile dyes, especially azo dyes, is complicated in aerobic conditions. However, in anaerobic conditions, the decolorization of textile dyes is better. The azo bond (—N⚌N—), the chromophore group in the chemical conformation of azo dyes, can be easily degraded in anaerobic conditions (Shahvali et al., 2000; Meehan et al., 2000; Guadie et al., 2018; Chandra et al., 2018). Among different microorganisms, bacteria can decompose dyes, especially azo dyes, due to faster growth, cheaper culture, and higher efficiency (Saba et al., 2013; Najme et al., 2015; Pan et al., 2017; Sharma et al., 2016). Iron-based nanoparticles have many applications in medicine. However, procedures of synthesis of FeNPs strongly affect their properties and limit their application. Recently, biological pathways for synthesizing iron nanoparticles have received more attention because the particles have suitable structural properties for biomedical disciplines. The mechanism of iron-based nanoparticle synthesis by microorganisms and its current limitations is studied (Arakha et al., 2015). FeNPs are used effectively in the purification of polluted waters (Zhang et al., 2011). Physical and chemical assays are effective methods for synthesizing iron nanoparticles (Kuhn et al., 2002; Karlsson et al., 2005). However, these methods are costly, poisonous, corrosive, and flammable chemicals (Shahwan et al., 2011). The size and form of nanoparticles are important in decolorization and can be controlled using different physical and chemical pathways (Naoki et al., 2008) (Sakulchaicharoen et al., 2010). Between the different technologies, nanotechnology has proven an unbelievable potential for remediation of wastewater and other environmental problems (Zare et al., 2013; Gupta et al., 2015). Environmentally friendly and low-cost nanomaterials with unique features have been produced for wastewater treatment, possible disinfection of industrial effluents, surface water, groundwater, and drinking water. Nanotechnology has been mentioned in various literature as one of the most developed sewage purification processes (Brumfiel, 2003; Theron et al., 2008). Fe2O3 is a cheap material for absorbing harmful metals due to its simple synthesis process. This material is environmentally friendly and can be utilized in a polluted environment with less risk of secondary pollution. The adsorption of various weighty metals on Fe2O nanoparticles relies on pH, temperature, and incubation time (Li et al., 2003). Iron nanoparticles are applied as antimicrobial agents and sewage treatment tools with low toxicity and increased irremediability. Research shows that iron oxide nanoparticles can damage nerve tissue by affecting glutathione levels and significantly increasing oxidation due to their high levels. Also, magnetic FeNPs can be utilized as a drug delivery medium (Prescott et al., 2002; Arunachalam et al., 2013) (Nazari and Jookar Kashi, 2020). It has recently been studied that stabilized nanoparticles have excellence over free nanoparticles. Calcium alginate seeds are current routine trapping cells that are cheap, non-toxic, and bioremediate. Also, trapped nanoparticles have high resistance compared to free particles. They are safe against toxins and can be used continuously (Bezbaruah et al., 2009) (Pantidos and Horsfall, 2014; Cho et al., 2015). This study aimed to isolate iron-producing bacteria from the Choghart mine, stabilize them in a sodium alginate bead and use them as catalysts. In this study, iron nanoparticles were produced for the first time from the novel bacterial strain S2. Also, the antibacterial activity and toxicity of iron nanoparticles were studied. Besides, stabilized FeNPs were utilized for dye elimination consumption.
2 Materials and methods
2.1 Dye and medium
The cultural media and materials utilized in this research were bought from Merck, Germany. Acid Red 88 dye was taken from Alvan Sabet Corporation, Iran.
2.2 Sample collection
All samples were prepared from a textile factory in Kashan, Iran, and the Choghart iron mine in Yazd, Iran, in November 2020. All samples were sent to the microbiology laboratory.
2.3 Microorganism isolation and identification
In this research, the bacterial strains were taken from the soil sample of the Choghart iron mine. The samples were diluted and cultured on nutrient agar with FeSO4 (20 and 50 mg/ml). Then, the plates were incubated at 30 οC for 24 h. We have subcultured colonies and, strain S2 was selected for further investigation. According to standard procedure, cell morphology, Gram-staining, oxidase, catalase, and KOH tests were carried for phenotypic characterization of strain S2 (Prescott et al., 2002). Marmur method was used to extract strain S2 chromosomal DNA (Marmur, 1961). Amplification of 16S rDNA genes was performed using general primers 27F and 1492R (Stackebrandt and Goodfellow, 1991; Turner et al., 1999). After purification and sequencing of PCR products, the sequences were sorted using Chromas Pro software from the official website https://www.technelysium.com.au. Then, the closest strains were identified on the Ezbiocloud website with a sequence of 16S rDNA similar to strain S2 (Yoon et al., 2017). Mega software was used to draw the strain S2 phylogeny tree with strains close to it (Milikli and Ramachandra Rao, 2012; Kumar et al., 2018). After finding the 16S rDNA sequence of close strains on the Ezbiocloud website, the phylogeny tree was plotted to align and calculate evolutionary distances with Bioedit software and the Neighbor-Joining, Maximum likelihood, and Maximum parsimony algorithms. The validity of tree branches was evaluated using the Bootstrap analysis algorithm with 1000 sampling times (Kimura, 1980; Tajima and Nei, 1984).
2.4 Bacterial synthesis of FeNPs
First, strain S2 was inoculated in 50 ml sterile nutrient broth. After 24 h incubation, the supernatant and biomass were separated using a centrifuge at 7,000 rpm for 15 min. Then, active supernatant and biomass were utilized to indicate synthesis of nanoparticles is biological. After that, the supernatant (1 ml) was dumped into 50 ml of Fe2(SO4)3 solution (2 mM) and was placed under a light bulb. The synthesis of FeNPs was evaluated using color change and UV–visible spectrophotometry (UV–Vis) analysis (300–800 nm). After 5 h, Fe2(SO4)3 solutions were centrifuged at 7000 rpm for 15 min. Then, nanoparticles were dried in an oven (UNB500, Memmert, Germany) for one day at 40–50 °C. After that, nanoparticles were placed in furnace at 450 °C for 4 h. FeNPs were identified by infrared spectroscopy (FTIR) (Magana 550, Nicolet, Germany), X-ray diffraction (XRD) (X Pert Pro, Philips, Netherlands), electron microscopy, and X-ray energy diffraction (SEM-EDX) (Mira3, Tescan Company, Czech). The iron oxide nanoparticle size was recorded using the formula 1 (Nautiyal and Shukla, 2018):
τ: The average size of crystallite (nm).
λ: The X-ray wavelength (nm).
β: Peak width at half the maximum height.
θ: Diffraction angle (°C).
2.5 Antibacterial effect
The antibacterial effect of FeNPs was evaluated by the agar diffusion assay on Escherichia coli (ATCC10536), Staphylococcus aureus (ATCC29737), Shigella dysentery (PTCC1188), Salmonella paratyphi-A serotype (ATCC5702), and Candida albicans (ATCC10231) according to clinical and laboratory standards institute (CLSI). The set of bacteria was prepared using the Iranian research organization for science and technology (IROST). The bacterial suspensions adjusted to 0.5 McFarland were inoculated on Mueller Hinton agar. Then, 20 µl of the FeNPs (40 mg/ml), Acid Red 88 dye (30 mg/ml), and decolorization product were loaded into 6 mm diameter wells. The Petri dishes were incubated overnight at 30 °C. The conclusions were indicated using examining the inhibition zone diameter.
2.6 Growth curve
The growth curve of strain S2 was drawn with UV–Vis using the following data:
First, strain S2 was inoculated in 5 ml of sterile nutrient broth and placed in a shaker incubator at 160 rpm at 30 °C for 24 h. Then 1 ml of the bacterial suspension was added into 100 ml of sterile broth nutrient medium and incubated at 30 °C. Thus, the absorption was read every 2 h at 600 nm.
2.7 Anticancer activity
The toxicity method was evaluated using the saltwater shrimp lethality test. A first, 0.1 g of Artemia salina eggs were exposed to light in an aquarium filled with salt water for 48 h by an aeration pump at room temperature. Then shrimp hatched and turned into larvae. Also, the saltwater pH inside the aquarium was set to 9 and all tests were performed in glass tubes consist of saltwater. Then, ten live larvae of saltwater shrimp were expanded with different concentrations (25, 250, 750, 1250, 1750, 2500 µg/ml) for FeNPs, Acid Red 88 dye decolorization product for 24 h. All tubes were prepared with 5 ml saline water and placed in a water bath under light at 30 °C. After 24 h, the amount of live and dead larvae any each tube was calculated. The evaluation was repeated three times to ensure the results. In this experiment, the control for each sample contained only 10 live larvae and 5 ml of saline water. The LC50 and mortality amount were calculated for various concentrations of FeNPs, Acid Red 88 dye, and decolorization product. Further, the results were compared with the control and indicated in the graphs. The mortality amount was assessed by formula 2 (Dashtizadeh and Jookar Kashi et al., 2021):
m: The average amount of dead larvae in the samples.
M: Mean amount of dead control larvae.
S: The average amount of live control larvae.
2.8 FeNPs/Ca-Alg beads
First, 20 mg/ml of FeNPs were mixed with 30 ml of sterile distilled water and was incubated in a shaker incubator for 20 min until wholly dispersed. Then, sterile distilled water with dispersed iron oxide nanoparticles was belended with 30 ml of 3% sodium alginate solution. After that, the solved solution was dropped dropwise to 40 ml of 3% CaCl2, producing beads in which FeNPs were trapped. FeNPs/Ca-Alg beads were utilized for Acid Red 88 dye decolorization.
2.9 Decolorization using FeNPs/Ca-Alg beads
First, 2 g of FeNPs/Ca-Alg beads were added into 20 ml of peptone-yeast medium with 50 g/l glucose and 25 g/l Acid Red 88 dye and incubated for 72 h at 30 °C. Then, 1 ml of the sample was centrifuged at 7000 rpm for 15 min at three intervals 24, 48, and 72 h. However, the discoloration was examined using UV–Vis. After each color change, FeNPs/Ca-Alg beads were washed and utilized again for decolorization. The test was repeated five times. Then, FeNPs/Ca-Alg beads were analyzed using SEM-EDX. The percentage of decolorization efficiency was evaluated using Formula 3 (Nazari and Jookar Kashi, 2020). Also, the metabolites after decolorization were prepared by ethyl acetate and evaluated using gas chromatography coupled with mass spectrometry. Finally, the dried powder of the sample was dissolved in ethyl acetate and injected into the device (6890, Agilent, USA).
2.10 Phytotoxicity test
The phytotoxicity of Acid Red 88 dye, the degraded dye metabolite were evaluated on the seed of Raphanus sativus L. First, the filter paper was placed inside plates, and sterilized in an autoclave (Abzar Teb Mahan, Iran). After that, the seed of R. sativus L. was sterilized with 70% alcohol and exposed to UV (Laminar Flow II (120RS, Jal tab, Iran) for half an hour. The seeds were sterilized in a plate with 70% alcohol for 1 min, distilled water for 10 min, and bleach 20% for 4 to 5 s. The seeds were placed in plates containing filter paper. 5 ml of the sample was added to the plates to determine toxicity. Control was performed with distilled water. The plates were placed at room temperature under sunlight. After 7 to 8 days, the difference is, the growth of seeds, the number of leaves, roots, and stems were measured. In this experiment, the conclusions were investigated using determining the relative root growth (RRG), relative seed germination (RSG), and germination index (GI) (Kebrom et al., 2019) as follows:
2.11 Resistance of strain S2 to extreme temperatures
For this test, strain S2, was inoculated into an Erlenmeyer flask including 50 ml of NB medium, and incubated in a shaker incubator (SI500, Stuart, UK). After 24 h, the supernatant and biomass were separated using a centrifuge (5810R, Eppendorf, Germany) at 4 οC and 7,000 rpm for 15 min. Then, bacterial cells and supernatant were autoclaved separately at 121 °C and 1.2 atmospheres for 15 min to investigate whether extracellular enzymes and the cell itself were resistant to high temperatures. Then the supernatant and biomass were used to produce iron nanoparticles separately.
3 Results and discussion
3.1 Investigation of iron tolerant bacteria
The soil samples were taken of the Choghart iron mine. Between the bacterial isolates obtained from the soil, an iron-tolerant strain S2 was picked to synthesize FeNPs. Also, this strain was resistant to high temperatures. Macroscopic (colony color), microscopic (morphological), and gram-staining were performed to identify bacterial strain S2. The strain was gram-positive with the white colony and coccobacillus morphology. Biochemical experiments performed to describe bacterial strain S2 are indicated in Table 1. Catalase, Oxidase, and KOH expriments were positive. Nucleotide level of strain S2 was utilized to investigate the phylogenetic connection between strain S2 and another isolate in the GenBank database (Fig. 1). The results showed that strain S2 relates to Bacillus genus belonging to JOTP01000061. Strain S2 was 99.68% similar to Bacillus zhangzhouensis DW5-4 (T), taken from the aquaculture water of a shrimp farm in Zhangzhous, China (Yang Liu et al., 2016). Further, the 16S rDNA gene sequence is linked to the gene bank (https://www.ncbi.nlm.nih.gov), and the accession number of strain S2 was obtained MZ668417. + positive control, ND (not determined).
Biochemical tests
Strain S2
B. zhangzhouensis DW5-4 T
Catalase
+
+
Oxidase
+
+
KOH
+
ND
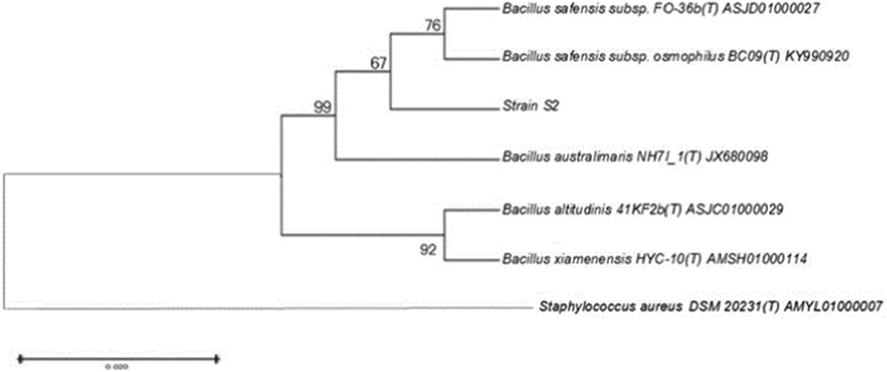
The phylogenetic tree shows the relationship between the strain S2 and other known homologous strains in the GenBank database.
3.2 Synthesis of FeNPs
The synthesis of FeNPs was evaluated with supernatant and biomass of strain S2. The supernatant and biomass were able to produce FeNPs separately. The ferrous sulfate solution changed from colorless to yellow, which confirmed the synthesis of FeNPs (Fig. 2. A-b). Fig. 2.A-a show UV–Vis spectra with the addition of the supernatant within 3 h. Also, strain S2 produced nanoparticles in high volumes (400 ml) (Fig. S1. B).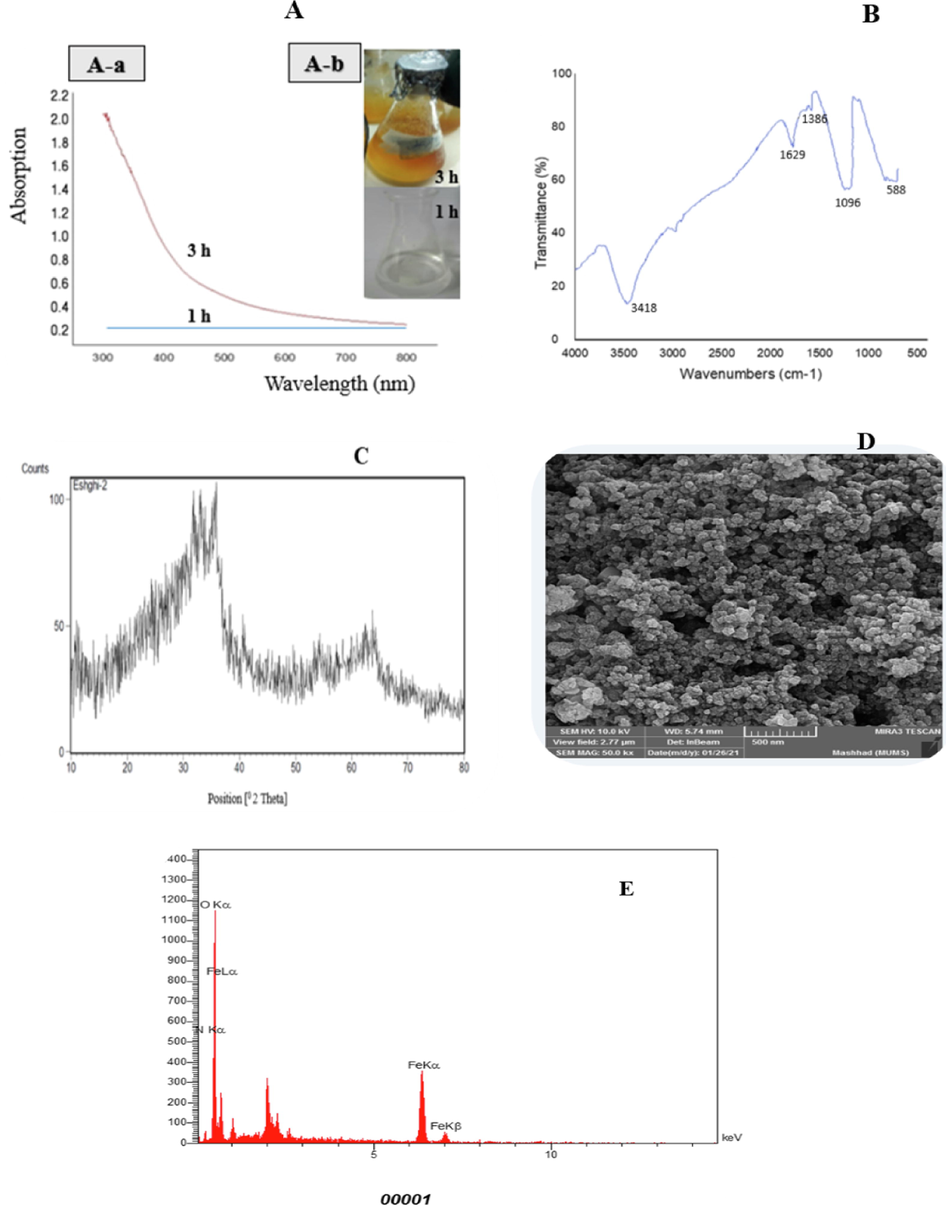
Spectrophotometric analysis (A), FTIR spectra (B), X-ray diffraction pattern (C), scanning electron microscopic images (D), and Energy dispersive X-ray spectrum (E) of FeNPs synthesized by strain S2.
3.3 Charactrization of FeNPs
Extracellular production of FeNPs was seen with discoloration to yellow. The color change of the FeSO4 solution to yellow was due to the reduction of iron ions and the production of nanoparticles, indicating the formation of iron nanoparticles. Also, the production of FeNPs was approved using UV–Vis at 300–800 nm. The most adsorption rate at 330 nm approved the make of nanoparticles (Fig. 2.A). Ultraviolet spectroscopy shows the production of FeNPs. Fadian Behbahani and Buazar (2019) synthesized magnetic iron oxide nanoparticles using marine Sargassum ilicifolium Seaweed. Their results indicated the color change of the Ferro ammonium sulfate solution from green to brown due to the reduction of iron ions and the production of iron nanoparticles.
The conclusions of the FTIR analysis are shown in Fig. 2. B. The adsorption band at 3418 cm−1 corresponds to the hydroxyl groups. The adsorption peak at 1629 cm−1 belonged to C⚌O. FTIR spectra showed that the carbonyl group belongs to amino acids and proteins, which have a great ability to bind to metal particles. The proteins play an impress in forming nanoparticles, preventing them from binding and stabilizing them in the environment. Generally, the FTIR results confirmed the presence of a protein layer with carbohydrates around the nanoparticles, which leads to the stability of the nanoparticles. The adsorption band at 1386 cm−1 shows the traction of C—H (Thakkar et al., 2010; Gurunathan et al., 2013; Mahdavi et al., 2013; Braihi, 2014; Gurunathan, 2019).
The XRD result was utilized to evaluate the structure of FeNPs. The three extreme peaks of the value of 2θ are 33, 35, and 63, corresponding to 100, 75, 40, respectively (Fig. 2.C). These results indicate the crystal structure of FeNPs. The average nanoparticle size calculated by the Scherer equation was 53.95 nm.
The SEM images proved the production of nanoparticles. The conclusions indicated that iron nanoparticles synthesized have a rhombus shape size of 20–70 nm (Fig. 2.D). FeNPs were deliberated using Digimizer software. FeNPs produced using supernatant were grouped depending on particles size scattering that minimum value was 10–20 nm and the maximum value was 30–40 nm (Fig. S2). Also, the EDX analysis of these particles indicates a peak in the iron region, which indicates the formation of FeNPs (Fig. 2.E). The reason for the intensification of the plasmon surface, an optical peak was observed in the 6/370KeV region, which corresponds to the adsorption of metal nanoparticles. Other peaks are carbon, oxygen, and nitrogen, which decide biomaterials on the nanoparticles. Also, the SEM images show the production of FeNPs. Moreover, the EDX spectrum proved the existence of a peak in the metal nanoparticles region.
3.4 Antimicrobial effect of FeNPs
The antibacterial activity of FeNPs was evaluated using the agar diffusion procedure against the tested strains. The FeNPs inhibited bacterial growth around wells as an inhibition zone. Fig. 3.A indicated that FeNPs have an antibacterial effect against tested bacterial strains. However, immobilized nanoparticles can be reused, so FeNPs/Ca-Alg bead effectively kills pathogenic bacteria. In a study, Torabian et al., 2019 perused the antibacterial effect of iron oxide nanoparticles synthesized by cytoplasmic extract of Lactobacillus casei. The inhibition zones of S. aureus at 100 and 1000 μg/ml concentrations were 12.03 ± 0.32 and 16 ± 0.5, respectively. In comparison, the nanoparticles are ineffective against E. coli.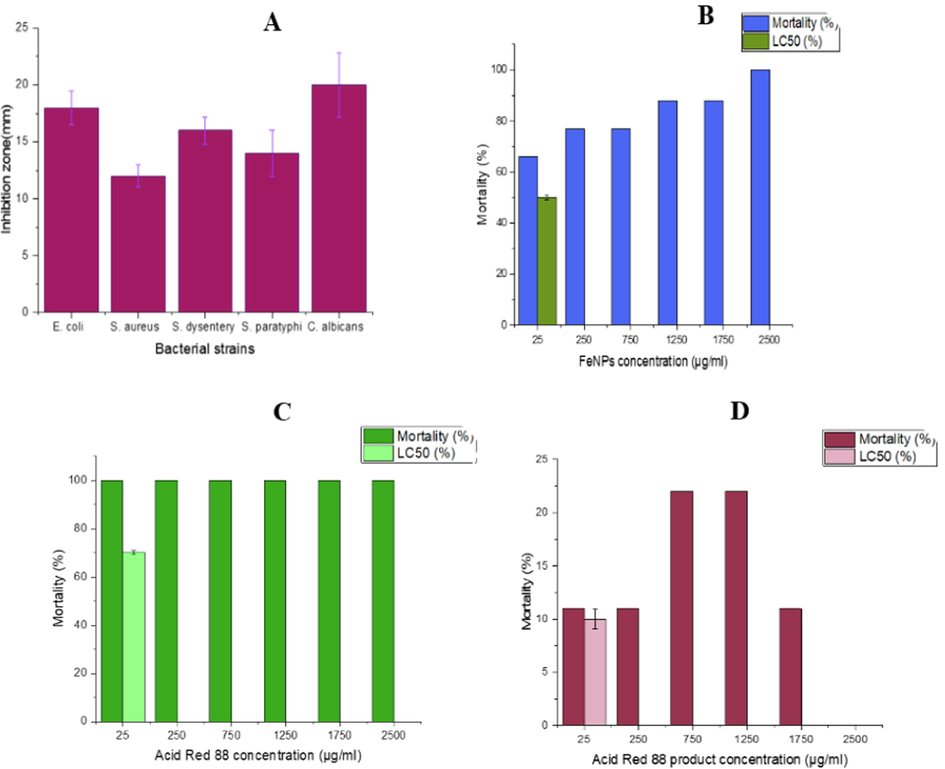
Antibacterial effect of FeNPs against standard strains (A), Biotoxicity assessment using A. salina after 24 h exposure to different concentrations of FeNPs (B), Acid Red 88 (C) and Acid Red 88 product (D).
3.5 Toxicity activity of FeNPs
3.5.1 Brine shrimp lethality test
In this research, the mortality assay of saltwater shrimp was utilized to assess the toxicity of FeNPs on living microorganisms. The conclusions showed notable mortality at 24 h (Fig. 3.B). More than 50% of the saltwater shrimp population was killed using FeNPs) 0.5 μg/ml(, which showed the high biotoxicity of the nanoparticles. Our results are similar to Negin Nazari et al. they applied the saltwater shrimp mortality assay to assess the toxicity of AgNPs on A. salina. Their results indicated that more than 50% of the A.salina was dead by 1 µg/ml AgNPs, which showed the toxicity of nanoparticles. In other studies, the toxicity of three nanoparticles, including chemical silver nanoparticles and biosynthetic silver nanoparticles prepared from aqueous extract of brown algae and green algae, were evaluated using brine shrimp lethality assay (BST) (Mashjoor et al. 2018). The nanoparticles at similar concentrations showed that chemical silver nanoparticles were up to 10 times more toxic than biosynthetic nanoparticles. Further, this study evaluated the toxicity of Acid Red 88 dye (Fig. 3.C) and its decolorization product (Fig. 3.D) by BST. The results indicated that the Acid Red 88 dye had very high toxicity, and the decolorization product had a very low toxicity. The decolorization product could not kill half of the shrimp. In a similar study, Satish Babu et al. used BST to measure Congo Red toxicity. Their results showed that the dye was very toxic, and the decolorization product by the new marine strain DTS26 was so few toxic (Satheesh Babua et al., 2015).
3.5.2 Germination seed for toxicity assay
In this study, radish seeds were also used to investigate the toxicity of FeNPs, Acid Red 88 dye, and its decolorization product. According to the data and formulas of section 2–10, the percentages of RRG and RSG for FeNPs were (0.18%, 18%), for Acid Red 88 dye were (0.16%, 16%), and for the product obtained from Acid Red 88 dye decolorization were (0.83%, 80%), respectively (Figure S3. A). Moreover, the GI for FeNPs, Acid Red 88 dye, decolorization product, and the test control (water) were 2.58%, 2.56%, 66.4%, and 100%, respectively (Fig. S3. B). The results show that the decolorization product of Acid Red 88 dye is very low toxicity, so it caused the growth of most seeds. In contrast, Acid Red 88 dye, and FeNPs due to their high toxicity, caused very low seed growth. In the control test, all seeds grew (100%). The size (length) of radish seeds was also measured using a ruler. The length of roots for control (H2O), FeNPs, Acid Red 88 dye, and decolorization products of Acid Red 88 were (3 cm, 0.5 cm, 0.5 cm, 2.5 cm), respectively (Fig. S3. C). Hashemi et al. 2018. reported the toxicity of zinc oxide nanoparticles upon the growth of Borago officinalis. According to their results, zinc oxide nanoparticles raised the root and stem length of plants. The reason for the grown root and stem of plants was the low toxicity of zinc oxide nanoparticles. Moreover, Chukowry et al. 2017 examined the cytotoxicity of azo dyes and their decolorization products by seed germination experiments. They concluded that the decolorization products of azo dyes were not toxic and did not inhibit germination.
3.6 Acid Red 88 dye decolorization using FeNPs/Ca-Alg beads
After the FeNPs/Ca-Alg beads were prepared, the beads were evaluated by SEM-EDX. The results indicated that FeNPs were carefully packaged on the inner surfaces of the alginate beads (Fig. 4.A to H). Also, these images show the porosity of the bead surface. The EDX spectrum indicates FeNPs/Ca-Alg beads, and the existence of FeNPs has been proved (Fig. 4.I). Also, the results show that the iron oxide nanoparticles are gotten stuck in Ca-Alg beads. These beads were utilized to decolorize Acid Red 88 dye in the next step. The conclusions indicated that Ca-Alg beads containing FeNPs decolorized and decomposed Acid Red 88 in 8 h with 100 % efficiency (Fig. 4.J). The reuse of FeNPs/Ca-Alg beads was investigated in five cycles. FeNPs/Ca-Alg beads were durable after any cycle. FeNPs/Ca-Alg beads could reuse to remove Acid Red 88 in five cycles with a 100% effect. However, the metabolites of decolorization were also evaluated using GC/MS analysis (Fig. 5). In the results obtained from this analysis, no mass was found related to the initial mass of the Acid Red 88 dye structure. This dye has a molecular weight of 400.38 g/mol, but the results do not show the presence of this dye after decolorization. Therefore, it can be concluded that this dye has decomposed and completely disappeared. The pathway of Acid Red 88 decomposition was proposed in Fig. 6. In 2015, Harshad Lade et al. used GC/MS analysis to recognize metabolites formed in Reactive Blue 172 dye decolorization using the bacterial strain HSL1. Their gas chromatography results indicated the presence of two peaks 4- (ethenylsulfonyl) aniline and 1-amino-1(4-aminophenyl) propane-2-one (Lade et al. 2015). Also, other researchers used GC/MS analysis to identify degraded metabolites during decolorization Procion Red-3B dye by Pseudomonas stutzeri SPM-1. Their results indicated that the presence of degraded dye metabolites (Bera and Tank 2021).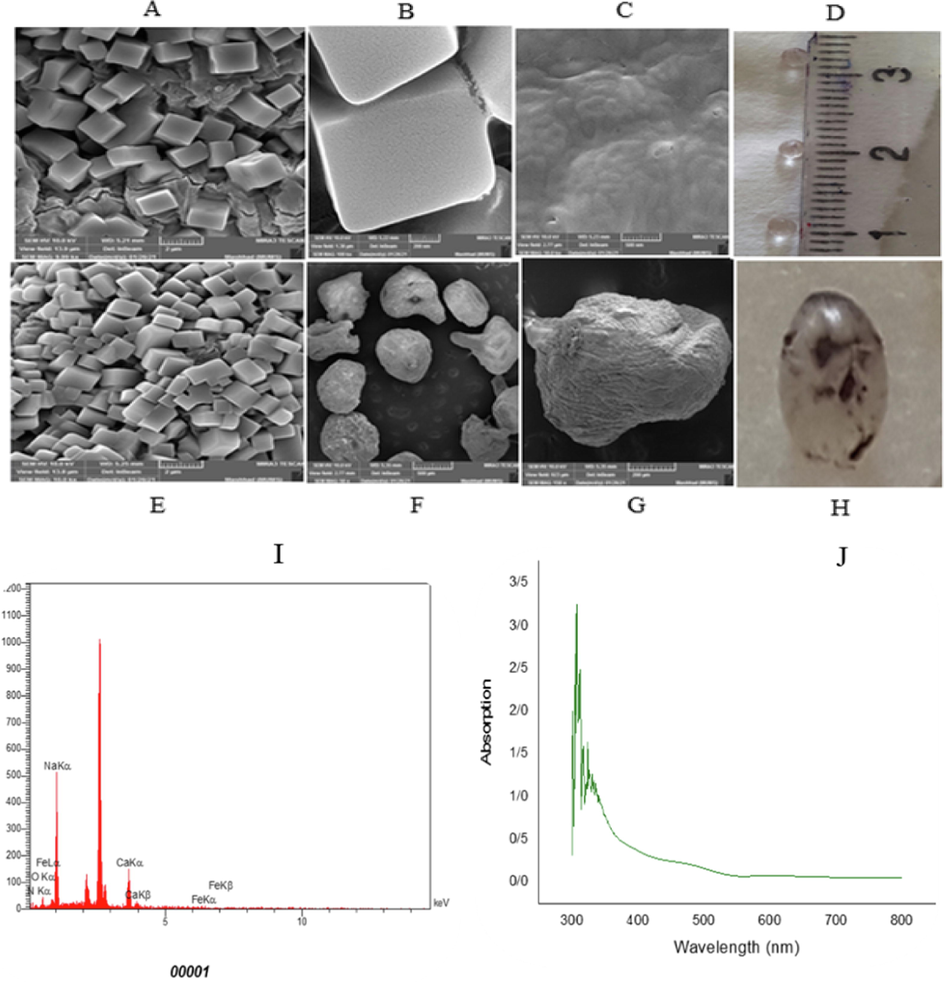
SEM images of outer and inner surface of FeNPs/Ca-Alg beads (A to G), Image of FeNPs/Ca-Alg beads (D, H), EDX spectra of FeNPs/Ca-Alg beads which confirms the presence of FeNPs (I), UV–Vis absorption spectra of decolorization of Acid Red 88 using FeNPs/Ca-Alg beads (J).

GC/MS analysis.
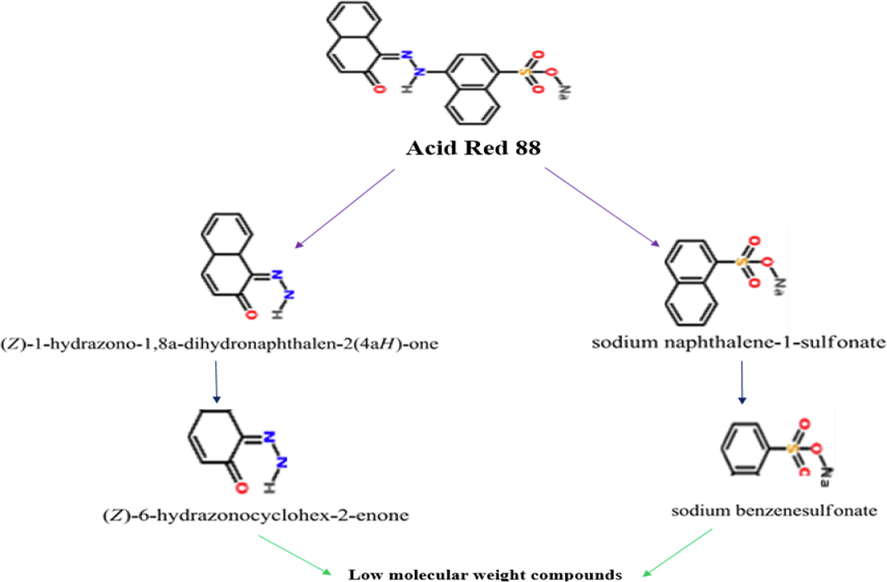
Acid Red 88 dye decomposition pathway.
3.7 Strain S2 growth curve
The growth curve of bacteria shows their growth pattern in four stages: delayed phase, logarithmic phase, standstill, and death phase. The bacteria cells multiply at the end of the active phase and the beginning of the stagnation phase of the growth curve. The bacterial growth curve was investigated using spectroscopy (Fig. S4). The bacteria reached the end of the logarithmic phase after 10 h. In other research, the growth curve of strain N23 isolated from urine samples was plotted. Their results showed that the bacterial strain reached the end of the logarithmic phase after 8 h)Fakher and Kashi, 2021).
3.8 Hyperthermophile strain S2
Bacterial strain S2 remained active after autoclaving at 121 °C and 1.2 atmospheres for 15 min. The results showed that strain S2 is resistant to high temperatures. This test showed that the extracellular enzymes were not damaged by autoclave because the autoclaved supernatant produced iron nanoparticles. Also, autoclaved bacterial cells were cultured in NA medium and incubated. After 24 h, strain S2 had grown and produced iron nanoparticles. Thus, strain S2 is hyperthermophile. In a similar report, Brock (1997) isolated strain 121 hyperthermophiles from a hot spring. Strain 121 was extremely resistant to high temperatures in an autoclave at 121 °C. It was able to double its population in 24 h. Also, the results indicated that strain 121 survived for 2 h at 130 °C but could not replicate until it was transported to a new growth culture at 103 °C.
4 Conclusion
A hyperthermophile B. zhangzhouensis strain S2 from the Ghoghart iron mine showed the capable synthesis of magnetic iron nanoparticles for the first time. The FeNPs was determined by UV–Vis spectroscopy, FTIR, XRD, SEM, and EDX analysis. FeNPs/Ca-Alg beads were utilized to decolorize Acid Red 88 dye for the first time. The GC/MS analysis confirmed that FeNPs /Ca-Alg beads could decolorize Acid Red 88 and completely decompose. Moreover, according to data from the antibacterial test, these nanoparticles have antibacterial properties. The phytotoxic test and BST showed that the dye toxicity decreased after decolorization. The results suggest that biologically synthesized iron nanoparticles can be used for wastewater.
Acknowledgement
We are grateful to University of Kashan for supporting this work.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics approval
This article does not contain any studies with human participants or animals performed by the authors.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: [Author 1 proclaims that the data stated in this article is from her master's thesis, submitted to Kashan University, North West, Isfahan, on 28th October 2021. The treatise is not presently published online or attainable by the public. The author has no conflict of interest to divulge. Author 2 proclaims that she has no conflict of interest.].
References
- Biosorption of Reactive Blue 49 dye under batch and continuous mode using a mixed biosorbent of macrofungus Agaricus bisporus and Thuja orientalis cones. Chem. Eng.. 2009;148(1):26-34.
- [CrossRef] [Google Scholar]
- Antimicrobial activity of iron oxide nanoparticle upon modulation of nanoparticle-bacteria interface. Sci. Rep.. 2015;5(1):14813.
- [CrossRef] [Google Scholar]
- One-step green synthesis and characterization of leaf extract-mediated biocompatible silver and gold nanoparticles from Memecylon umbellatum. Int. J. Nanomed.. 2013;8:1307-1315.
- [CrossRef] [Google Scholar]
- Microbial degradation of Procion Red by Pseudomonas stutzeri. Sci. Rep.. 2021;11(1):3075.
- [CrossRef] [Google Scholar]
- Entrapment of iron nanoparticles in calcium alginate beads for groundwater remediation applications. J. Hazard. Mater.. 2009;166(2–3):1339-1343.
- [CrossRef] [Google Scholar]
- Int. J. Mater. Sci. Appl.. 2014;3:363.
- The Value of Basic Research: Discovery of 117termus aquaticus and Other. Extreme Thermophiles. 1997;146(4):1207-1210.
- [CrossRef] [Google Scholar]
- Evaluation of molasses-melanoidin decolourisation by potential bacterial consortium discharged in distillery effluent. 3 Biotech.. 2018;8(4):187.
- [CrossRef] [Google Scholar]
- Reduction of nitrate in groundwater by Fe (0)/Magnetite nanoparticles entrapped in Ca-Alginate beads. Water, Air, Soil Pollut.. 2015;226(7):206.
- [Google Scholar]
- Bacillus algicola decolourises more than 95% of some textile azo dyes. Environ. Chem. Lett.. 2017;15(3):531-536.
- [CrossRef] [Google Scholar]
- Phytosynthesis of copper nanoparticles using Prunus mahaleb L. and its biological activity. Mater. Today Commun.. 2021;27(6):102456.
- [CrossRef] [Google Scholar]
- Green Synthesis and Characterization of Magnetic Iron Oxide Nanoparticles Using Marine Sargassum ilicifolium. Seaweed.. 2019;18(1):25-32.
- [Google Scholar]
- Microbial Synthesized Ag/AgCl Nanoparticles Using Staphylococcus pasteuri sp. nov., ZAR1: Antimutagenicity, Antimicrobial Agent. J. Inorganic Organometallic Polym. Mater.. 2021;31(2):1688-1703.
- [CrossRef] [Google Scholar]
- Halomonas sp strain A55, a novel dye decolorizing bacteriumfromdye-uncontaminated Rift Valley Soda lake. Chemosphere. 2018;206:59-69.
- [CrossRef] [Google Scholar]
- Nanoparticles as Adsorbent; A Positive Approach for Removal of Noxious Metal Ions: A Review. Sci. Technol. Develop.. 2015;34:195.
- [Google Scholar]
- Rapid biological synthesis of silver nanoparticles and their enhanced antibacterial effects against Escherichia fergusonii and Streptococcus mutans. Arab. J. Chem.. 2019;12(2):168-180.
- [CrossRef] [Google Scholar]
- Green synthesis of silver nanoparticles using Ganoderma neo-japonicum Imazeki: a potential cytotoxic agent against breast cancer cells. Int. J. Nanomed.. 2013;8:4399-4413.
- [CrossRef] [Google Scholar]
- Hashemi, S., Nadernejad, N., Pourseyedi, S., Asrar, Z., 2018. Investigation of toxicity of zinc oxide nanoparticles synthesized by olive extract on growth and pigments in Borago officinalis. 8(30), 303–315.
- Size-controlled nanoparticles by thermal cracking of iron pentacarbonyl. Appl. Phys. A: Mater. Sci. Process.. 2005;80:1579-1583.
- [Google Scholar]
- Evaluation of phytotoxicity of three organic amendments to collard greens using the seed germination bioassay. Environ. Sci. Pollut. Res.. 2019;26:5454-5462.
- [CrossRef] [Google Scholar]
- A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol.. 1980;16(2):111-120.
- [CrossRef] [Google Scholar]
- Structural and magnetic properties of core-shell iron-iron oxide nanoparticles. J Phys Condens Matter.. 2002;14(49):13551.
- [CrossRef] [Google Scholar]
- MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.. 2018;35(6):1547-1549.
- [CrossRef] [Google Scholar]
- Lade Harshad, Govindwar Sanjay, Paul Diby, 2015. Low-Cost Biodegradation and Detoxification of Textile Azo Dye C.I. Reactive Blue 172 by Providencia rettgeri Strain HSL1. 4, 1-10. https://doi.org/10.1155/2015/894109.
- Adsorption of cadmium (II) from aqueous solution by surface oxidized carbon nanotubes. Carbon. 2003;41(5):1057-1062.
- [CrossRef] [Google Scholar]
- Bacillus zhangzhouensis sp. nov. and Bacillus australimaris sp. nov. Int. J. Syst. Evol. Microbiol.. 2016;66(3):1193-1199.
- [CrossRef] [Google Scholar]
- Synthesis, Surface Modification and Characterisation of Biocompatible Magnetic Iron Oxide Nanoparticles for Biomedical Applications. Molecules. 2013;18(7):7533-7548.
- [CrossRef] [Google Scholar]
- A review onbacterial degradation of textile dyes. J. Chem. Chem. Sci.. 2013;3(3):201-212.
- [Google Scholar]
- A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol.. 1961;3(2):208-218.
- [CrossRef] [Google Scholar]
- Comparative Toxicity Assessment of Chemical Nanosilver and Biosynthetic Silver Nanoparticles Produced by Marine Macroalgae from the Persian Gulf in Biomarker. Artemia nauplii.. 2018;9(3):465-472.
- [CrossRef] [Google Scholar]
- Marchant R, Decolorization of Remazol Black B using a thermotolerant yeast, Kluyveromyces marxianus IMB3. Environ Int.. 2000;26(1–2):75-79.
- [CrossRef] [Google Scholar]
- Molecular characterization and optimization of bacteria in azo dye degradation. Int. J. Int Sci. Inn. Tech. Sec. B.. 2012;1(5):32-39.
- [Google Scholar]
- Biodecolorization of reactive yellow-2 by Serratia sp RN34 isolated from textile wastewater. Water Environ. Res.. 2015;87(12):65-75.
- [CrossRef] [Google Scholar]
- Naoki T., Macromol Symp., et al., 2008. Decolorization of synthetic wastewaters by nickel oxide nanoparticle, 1(1), 25.
- Silver nanoparticles catalyzed reductive decolorization of spent dye bath containing acid dye and its reuse in dyeing. J. Water Process Eng.. 2018;22:276-285.
- [CrossRef] [Google Scholar]
- A novel microbial synthesis of silver nanoparticles: Its bioactivity, Ag/ Ca-Alg beads as an effective catalyst for decolorization Disperse Blue 183 from textile industry effluent. Sep. Purif. Technol.. 2020;259
- [CrossRef] [Google Scholar]
- Decolorization pathways of anthraquinone dye Disperse Blue 2BLN by Aspergillus sp. XJ-2 CGMCC12963. Bioengineered.. 2017;8(5):630-641.
- [CrossRef] [Google Scholar]
- Biological synthesis of metallic nanoparticles by bacteria, fungi and plants. J. Nanomed. Nanotechnol.. 2014;5(5):1000233.
- [CrossRef] [Google Scholar]
- Prescott, Harley, Klein, 2002. Microbiology. 5th ed., The McGraw−Hill Companies. Boston, pp. 978–981. https://doi.org/10.4236/oalib.1103783.
- Development ofmicrobial consortium for the biodegradation andbiodecolorization of textile effluents. J. Urban Environ. Eng.. 2012;6(1):36-41.
- [Google Scholar]
- Biodegradation of mixed textile dyes by bacterial strainisolated from dye waste effluent. Res. J. Environ. Toxicol.. 2011;5(2):97-107.
- [CrossRef] [Google Scholar]
- Reactive black-5 azo dye treatment in suspended and attach growth sequencing batch bioreactor using different co-substrates. Int. Biodeterior. Biodegrad.. 2013;85:556-562.
- [CrossRef] [Google Scholar]
- J. Contam. Hydrol.. 2010;118(3–4):117-127.
- Detoxification and color removal of Congo red by a novel Dietzia sp. (DTS26) – A microcosm approach. Ecotoxicol. Environ. Saf.. 2015;14:52-60.
- [Google Scholar]
- Effect of environmental parameters on decolorization of textile wastewater using Phanerochaete chrysosporium. Bioproc Eng.. 2000;23(6):721-726.
- [CrossRef] [Google Scholar]
- Green synthesis of iron nanoparticles and their application as a Fenton-like catalyst for the degradation of aqueous cationic and anionic dyes. Chem. Eng. J.. 2011;172(1):258-266.
- [CrossRef] [Google Scholar]
- Decolorization of azo dye methyl red by suspended and co-immobilized bacterial cells with mediators anthraquinone-2,6-disulfonate and Fe3O4 nanoparticles. Int. Biodeterior. Biodegrad.. 2016;112:88-97.
- [Google Scholar]
- Nucleic acid techniques in bacterial systematics. Wiley.. 1991;31(6):479-480.
- [CrossRef] [Google Scholar]
- Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol.. 1984;1(3):269-285.
- [CrossRef] [Google Scholar]
- Biological synthesis of metallic nanoparticles, Nanomedicine Nanotechnology. Biol. Med.. 2010;6(2):257-262.
- [CrossRef] [Google Scholar]
- Nanotechnology and water treatment: Applications and emerging opportunities. Crit. Rev. Microbiol.. 2008;34(1):43-69.
- [CrossRef] [Google Scholar]
- Evaluating Antibacterial Effect of Green Synthesis Oxide Iron Nanoparticles Using Cytoplasmic Extract of Lactobacillus casei. J. Babol Univ. Medical Sci.. 2019;21(1):237-241.
- [Google Scholar]
- Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis 1. J. Eukaryot. Microbiol.. 1999;46(4):327-338.
- [CrossRef] [Google Scholar]
- Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol.. 2017;67(5):1613-1617.
- [CrossRef] [Google Scholar]
- Studies of ab initio and Monte Carlo simulation on interaction of fluorouracil anticancer drug with carbon nanotube. J. Nanostructure Chem.. 2013;3(1):1-8.
- [Google Scholar]
- Kaolinite-supported nanoscale zerovalent iron for removal of Pb2+ from aqueous solution: reactivity, characterization and mechanism. Water Res.. 2011;45(11):3481-3488.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.104032.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







