Translate this page into:
Butea monosperma seed extract mediated biosynthesis of ZnO NPs and their antibacterial, antibiofilm and anti-quorum sensing potentialities
⁎Corresponding authors. syedmicro72@gmail.com (Syed Ghazanfar Ali), aamtrody@qu.edu.sa (Ahmad Almatroudi), sfadil@ksu.edu.sa (Syed Farooq Adil)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Nanoparticles (NPs) prepared through safer, eco-friendly and natural-based products can be used various applications including antimicrobial and biomedical applications. Quorum sensing (QS) is a complex system for intercellular communication that regulates the expression of many virulence determinants, in the opportunistic Pseudomonas aeruginosa. Thus, the disruption of QS-mediated communication system could be an alternative strategy to combat infection caused by drug resistant P. aeruginosa. In this study, zinc oxide nanoparticles (ZnO NPs) were synthesized by using seed extract of Butea monosperma and further characterized by XRD, SEM and TEM. The biosynthesized ZnO NPs were then subjected to the investigation of their antibacterial, antibiofilm activities as well as the anti-quorum sensing potentialities for biomedical applications. The minimum inhibitory concentration (MIC) value against P. aeruginosa was found 1600 µg/ml, and it was found that ZnO NPs effectively reduced virulence factors Viz. swarming and swimming of P. aeruginosa by 62.8 and 45%, respectively. Exopolysaccharides production was also affected by ZnO NPs which indicate the disruption of biofilm formation. Further, the in silico docking analysis displayed that ZnO NPs efficiently interacted within QS (Las/Rhl) mechanism of P. aeruginosa by binding to the catalytic cleft of LasI synthase (Gly-116-ZnO = 2.8 Å), RhlI synthase (Gly-180-ZnO = 3.7 Å) and transcription receptor protein LasR receptor (Asn-141-ZnO = 2.9 Å), RhlR receptor (Tyr-72-ZnO = 2.6 Å). Thus, it was concluded that the effective interaction of ZnO NPs with Las and Rhl system led to the inactivation of autoinducers such as N-acyl homoserine lactones (AHL) molecules which finally led to the inactivation of QS and down regulation of virulence determinants. Consequently, the present in vitro and in silico results obtained suggest that the synthesized ZnO NPs can be efficiently used as potent antimicrobial agents to prevent the colonization, biofilm formation as well as anti-QS-mediated determinants caused by pathogenic drug resistant bacteria.
Keywords
ZnO NPs
Biofilm
Antimicrobial
In silico
Pseudomonas aeruginosa
Quorum sensing
Las/Rhl systems
Virulence determinants
1 Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that causes a number of diseases such as a respiratory tract infection, nosocomial infections, urinary tract infection, surgical wound infection, and secondary infection in HIV infected patients (Palmer et al., 2005). Quorum sensing (QS) is a complex system for intercellular communication that regulates the expression of many virulence determinants (such as viz. pyocyanin, protease, toxins, swarming, swimming, and biofilm formation), particularly in the opportunistic human pathogen P. aeruginosa. The mechanism of pathogenicity involves the production, detection, and response to extracellular signaling molecules which are called autoinducers (Longo et al., 2013). The autoinducer molecule i.e., acyl homoserine lactone (AHL) present in P. aeruginosa, is responsible for the switching on of cascade of the gene associated with antibiotic resistance, virulence, and biofilm formation (Pearson et al., 1997; Pesci et al., 1999; Van Delden and Iglewski, 1998). Nanoparticles could be used as an effective remedy against bacterial infection, as the bacteria do not develop resistance against nanoparticles (Mühling et al., 2009).
P. aeruginosa employs two QS systems for maintaining pathogenicity and multidrug resistance and these systems involve LasI/LasR and RhlI/RhlR which follow a series of steps including generation of autoinducers molecules, intercellular communication and the expression of virulence determinants. The genes encoded by the two systems are primarily LasI and RhlI genes (De Kievit et al., 2001; Koch et al., 2005). Although, Pseudomonas quinolone signal (PQS) system is the third type of system that also affects cells by altering transcriptional profiles of genes and binds to the previously unrecognized protein partners. Despite these function the precise role of PQS in vitro and in vivo is poorly understood (Lin et al., 2018). The LasI QS system is generally activated by 3-oxo-C-12 homoserine lactone (HSL) and RhlI is activated by C4-HSL, and the gene LasI and RhlI synthase synthesize 3-oxo-C12-HSL and C4-HSL, respectively. When a certain threshold concentration of 3-oxo C-12 HSL and C4-HSL is achieved, then they bind to their regulators i.e., LasR and RhlR and forms LasR-3-oxo-C-12-HSL and RhlR-C4-HSL complex, respectively. These LasR and RhlR complex, further, activates the expression of virulence genes (Chelvam et al., 2014). Hence, an alternative strategy to fight against P. aeruginosa infection must involve the interference with the QS system, since these molecules regulate virulence determinants (Alvarez et al., 2012).
In silico approach is the latest approach based on the computational analysis and nowadays used for the drug screening and discovery program. This approach also allows us to predict the most probably binding mode of candidate hits onto the selected target which further provides a molecular basis for their optimization in terms of binding affinity (Reuter et al., 2016). In silico-based approaches have shown the anti-QS nature of silver nanoparticles (Ali et al., 2017; Vyshnava et al., 2016) but very few reports are available on anti-QS strategy of nanoparticles using molecular docking approach.
Based on the literature reports about the inability of the pathogenic bacterial strains to develop resistance towards the NPs and in continuation of the efforts of our group to synthesize nanoparticles for various applications(Adil et al., 2015; Anandan et al., 2019; Shobha et al., 2020; Sumanth et al., 2020; Khan et al., 2016; Khan et al., 2014; Khan et al., 2018; Lakshmeesha et al., 2020; Prasad et al., 2020), the objective of the present work was (i) biosynthesis of ZnO NPs using seed extract of Butea monosperma, (ii) characterization of the biosynthesized ZnO NPs by XRD, SEM and TEM, (iii) In vitro investigation of antibacterial, antibiofilm and anti-QS potentialities of the biosynthesized ZnO NPs, (iv) In silico and molecular docking analysis of interaction of ZnO NPs with QS-controlled proteins LasI/RhlI AHL synthase and (v) In silico and molecular docking analysis of interaction of the biosynthesized ZnO NPs with QS-controlled transcriptional regulatory proteins LasR/RhlR.
2 Materials and methods
2.1 Biosynthesis ZnO NPs
ZnO NPs were synthesized as method described by Ali et al. (Ali et al., 2020). Briefly, 40 ml Butea monosperma seed extract was mixed with 1 mM Zinc nitrate hexahydrate (HiMedia, Mumbai, India) solution. The cream colored precipitate was obtained after heating at 60 °C for 3–4 h, which was further centrifuged and oven-dried at 45 °C for 24–48 h and the obtained ZnO nanopowder was used for further study.
2.2 Characterization of biosynthesized ZnO NPs
2.2.1 XRD analysis
The crystalline behavior of biosynthesized ZnO NPs was analyzed by XRD analysis using Bruker D8 Diffractometer (Berlin, Germany) and the particle size (D) calculated using Scherer equation: where λ is the wavelength of X-ray, B is the broadening of the diffraction line measured half of its maximum intensity and θ is the Bragg’s angle.
2.2.2 SEM and TEM analysis of biosynthesized ZnO NPs
The morphology, shape, size and particles distribution of biosynthesized ZnO NPs were analyzed by SEM (JSM 6510 LV) and TEM (JEM 2100) manufactured by JEOL from Japan using protocols used in our previous study(Ali et al., 2020).
2.3 Evaluation of minimum inhibitory concentration (MIC) of ZnO NPs
The Minimum inhibitory concentration (MIC) of ZnO NPs against P. aeruginosa was determined using microbroth dilution method in a 96-wells microtiter culture plate (Ali et al., 2020). Briefly, P. aeruginosa were cultured on nutrient agar (HiMedia, Mumbia, India) plates at 37 °C for overnight and then next day single colony was inoculated into the nutrient broth (HiMedia, Mumbia, India) for 5–6 h at 37 °C on rotor shaker with 100 rpm to get the bacterial growth of 105 to 106 CFU/ml. The ZnO NPs were serially diluted with different concentrations (3200–100 μg/mL). Then, 20 μl of P. aeruginosa culture was added into 180 μl nutrient broth and then kept in incubator at 37 °C for 24 h. MIC is defined as the lowest concentration of antimicrobial agents that visually inhibit bacterial growth by 99%.
2.4 Inhibition of virulence
2.4.1 Qualitative assessment of antibiofilm potential of ZnONPs by Congo red agar method
Antibiofilm potential of nanoparticles was assessed using Congo red agar (CRA) method. Briefly, BHI (37 g/L), sucrose (50 g/L), and bacteriological agar (10 g/L) (HiMedia, Mumbia, India) were autoclaved and concentrated Congo red solution (0.8 g/L) (HiMedia, Mumbia, India) was also autoclaved separately. After autoclave media and congo red were allowed to cool to a temperature about 50–55 °C, and after cooling congo red was mixed with BHI (HiMedia, Mumbia, India) media. The ZnO NPs with the desired concentration were also added into the media and poured into the plates. Control plates were not inoculated with nanoparticles. Overnight grown culture of P. aeruginosa was inoculated on to the media with or without nanoparticles and incubated at 37 °C for 24 h.
2.4.2 Quantitative assessment of antibiofilm potential of ZnO NPs by crystal violet assay
Biofilm formation was evaluated as described by O’Toole and Kolter (1998) with slight modification. Briefly, 2 ml eppendrof tubes were inoculated by 100 µl (1x107 CFU/mL) of the mid-exponential culture of P. aeruginosa with 300 µg/mL of ZnO NPs and other tube was inoculated with P. aeruginosa without nanoparticles. The tubes were then incubated for 24 h at 37 °C. After 24 h of incubation, tubes were washed with distilled water, air dried and then stained with 0.1%w/v crystal violet for 30 min, after that, crystal violet was removed and the tubes were again washed with distilled water three times, dried and filled with absolute ethanol and then optical density were recorded at 595 nm.
2.4.3 Assessment of antibiofilm potential of ZnO NPs by confocal laser scanning microscopy (CLSM)
Antibiofilm activity of ZnO NPs was assessed using CLSM as described by Ansari et al. (2014). Briefly, in a 12 well microtitre plate, 4 ml media (BHI + 5% sucrose) was added along with glass coverslip. In one of the well ZnO NPs (300 µg/mL) was added and other well was not added with nanoparticles. Mid-exponential overnight grown culture of P. aeruginosa (100 µl) was added in each well. Plate was left for incubation for 24 h at 37 °C. After the incubation of 24 h, glass cover slips were removed and carefully washed with sterile PBS and then incubated with 50 µg/mL of concanavalin- A conjugated fluorescein isothiocyanate (ConA-FITC, Sigma–Aldrich) for 15 min at room temperature. The Con A-FITC was excited at 495 nm and monitored at an emission wave length of 525 nm. Intact biofilms were examined using a Fluoview FV1000 Espectral Olympus CSLM (Olympus Latin America, Miami, FL, USA) equipped with a UPlanSApo 100x/1.40 oil UIS2 Olympus oil immersion lens.
2.4.4 Effect of nanoparticles on swarming motility of P. aeruginosa
Swarming motility was assessed as described by Kalai-chelvam et al. (Chelvam et al., 2014). Briefly, semi-solid agar (HiMedia, Mumbia, India) was prepared with or without nanoparticles using nutrient broth, bacteriological agar (0.5%), and 0.5% glucose (HiMedia, Mumbia, India). After cooling the nanoparticles with desired concentration were added to the media and were poured in the plates. Control plates were not amended with nanoparticles. Overnight grown culture of P. aeruginosa was spot inoculated on both the plates (with and without nanoparticles) and incubated at 37 °C for 24 h.
2.4.5 Effect of nanoparticles on swimming motility of P. aeruginosa
Swimming motility was also performed as described by Kalai-chelvam et al. (Chelvam et al., 2014). Briefly, semi-solid agar media was prepared with or without nanoparticles using nutrient broth, bacteriological agar (0.25%), and 0.5% glucose. The media was autoclaved and cooled before the addition of nanoparticles. The media was then allowed to solidify after the addition of nanoparticles in desired concentration by keeping the plates remain in laminar for 24–48 h. Plates without nanoparticles were treated as a control. Spot inoculation of an overnight grown culture of P. aeruginosa was done on the plates (with and without nanoparticles) and kept for incubation at 37 °C for 24 h.
2.5 In silico analysis
2.5.1 Preparation of 3D structure of Las synthase and Las receptor molecules
The 3D structure of LasI Synthase (PDB ID: 1RO5) and transcriptional activator protein LasR (PDB ID: 2UV0) were retrieved from Protein Data Bank. The 3D crystal structure of RhlI Synthase and its regulatory protein RhlR were not available in the Protein Databank. RhlI and RhlR synthase were modelled using SWISS-MODEL homology method. SWISS-MODEL integrated with PROMOD-II tool was used for the 3D model building based on target template alignment (Guex et al., 2009; Šali and Blundell, 1993). To estimate the overall quality of modelled 3D structure of RhlI Synthase and RhlR, QMEAN scoring function was analysed by SWISS MODEL (Benkert et al., 2011). Further, RAMGAGE Ramachandran Plot analysis server was used to validate the model structure (Prisant et al., 2003).
2.5.2 Preparation of 3D structure of ZnO for docking analysis
The .mol file of ZnO was downloaded from Protein Data Bank and converted into .pdb files. The minimization of ligand was done by using Chimera version 1.10 (Pettersen et al., 2004).
2.5.3 Prediction of active site of Las and Rhl systems
The binding affinity and identification of active binding sites of ZnO to Las and Rhl was done by a 3D ligand site developed by the structural bioinformatics groups, Imperial College London (Wass et al., 2010).
2.5.4 Molecular docking analysis
The interaction of ZnO to LasI/R and RhlI/R synthase was explored through the Patchdock tool (Ali et al., 2017; Schneidman-Duhovny et al., 2005). The selected docked file was subjected to the post-docking 3D simulation using discovery studio visualize 4.5 and PyMol 3D visualization software (Ali et al., 2017).
3 Results and discussions
3.1 Characterization of biosynthesized ZnO NPs
3.1.1 XRD analysis
The biosynthesized ZnO NPs were characterized by XRD which revealed the crystalline nature of ZnO NPs. The diffraction peaks obtained at 31.8°, 34.5°, 36.2°, 47.7°, 56.6°, 62.8°, 66.2°, 68.0°, 69.1°, 72.6° and 77.2° were indexed to 100, 002, 101, 102, 110, 103, 200, 112, 201, 004 and 202, respectively. The diffraction peaks were indexed according to the JCPDS No. 36–1451 and no characteristic peaks of other impurities and intermediate phases were observed. The XRD pattern shows the hexagonal phase of zinc oxide i.e., wurtzite structure. The sharp diffraction peaks suggest that the biosynthesized ZnO NPs were crystalline in nature and the average crystal size of ZnO NPs was found to be 24.5 nm (Fig. 1).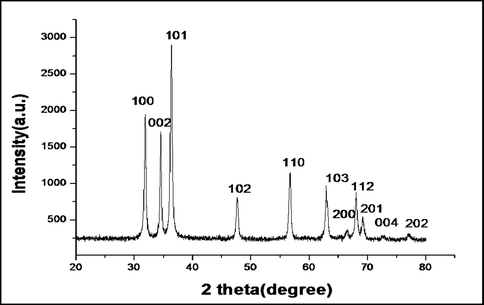
XRD pattern of biosynthesized ZnO nanoparticles.
3.1.2 SEM and TEM analysis
The morphologies of ZnO NPs were revealed by SEM analysis. Figure (2a) represents the SEM of synthesized ZnO NPs with slight aggregation and clumping. TEM analysis depicted the morphology and size of nanoparticles. TEM image (Fig. 2b) shows that the nanoparticles were hexagonal, rectangular and some were spherical in shape. The average particle size of ZnO NPs was 24.62 nm.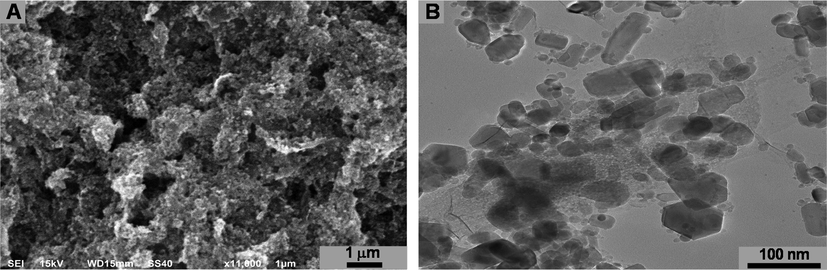
SEM (A) and TEM (B) micrograph of biosynthesized ZnO NPs.
3.2 Evaluation of antibacterial activity (MIC)
In the present study, the MICs of ZnO NPs against P. aeruginosa was evaluated using two-fold serial microbroth dilution method and the MIC value of ZnO NPs was 1600 µg/mL.
3.3 Inhibition of virulence factors
3.3.1 Swarming, swimming, and biofilm
Motility is a QS dependent virulence function that has a key role in biofilm formation since it helps the cell in the surface attachment (Pratt and Kolter, 1998). Swarming is a kind of social strategy that coordinates the movement of bacteria across the semisolid surface (Köhler et al., 2000). The motility factors viz. swarming and swimming of P. aeruginosa were effectively decreased by ZnO NPs (Fig. 3). Swarming motility decreased by 62.80% whereas swimming motility decreased by 45% upon treatment with 300 µg/mL of ZnO NPs (Fig. 4). Similar concentration of ZnO NPs also decreased the biofilm formation of PAO1 on Congo red agar, decrease in the secretion of exopolysaccharide i.e. absence of black crystalline colonies were observed (Fig. 5A & B). The quantitative crystal violet assay shows that ZnO NPs effectively decreased the biofilm formation by 57.08% at 300 µg/mL (Fig. 5C). Further, CLSM also showed the decrease in biofilm formation since the biofilm forming cells upon treatment with ZnO NPs did not remain intact onto the surface (Fig. 6). Our results are also in agreement with the previous finding of Khan et al. (2020) who have shown through CLSM that Zinc spikes effectively decreased the biofilm formation. Khan et al. (2020) showed maximum 58% and 85% reduction in biofilm at 200 µg/ml of different zinc nanostructures. Lee et al. (2014) also showed the similar results of decrease in biofilm formation using ZnO NPs. Interestingly, the condition of swarming mimic with the mucous layers that overlay epithelial surfaces, such as lungs which are major sites of infection by P. aeruginosa in patients of cystic fibrosis (Hutchison and Govan, 1999).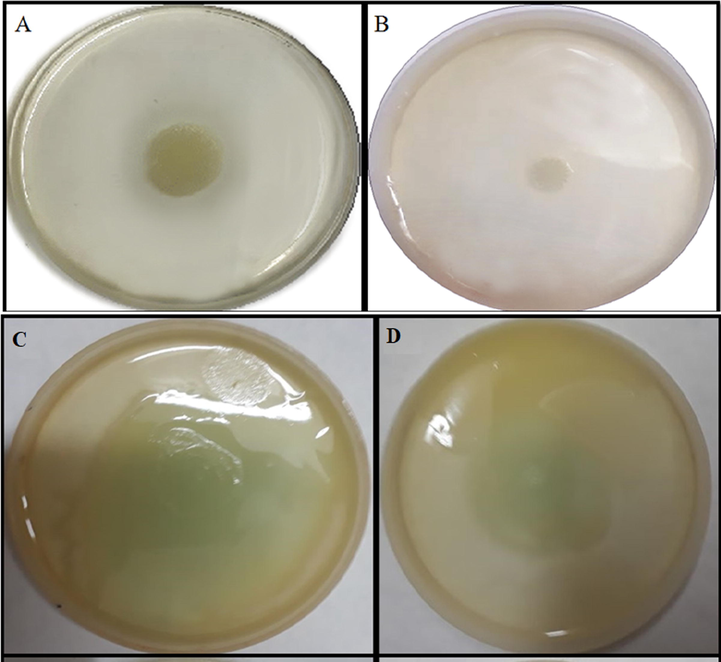
Effect of ZnO nanoparticles on swarming (A&B) & swimming (C&D) motility of P. aeruginosa. A & C are control images whereas B & D are nanoparticles amended with 300 µg/mL of ZnO NPs.
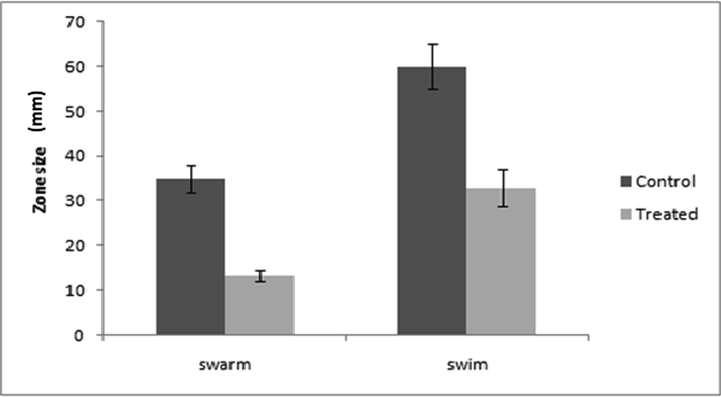
Bar graphs of P. aeruginosa PAO1 representing swarming and swimming motilities before and after treatment with ZnO NPs (300 µg/mL). Swarming and swimming expressed as diameter of swarm and swim in mm. Error bars are representative of standard deviation.
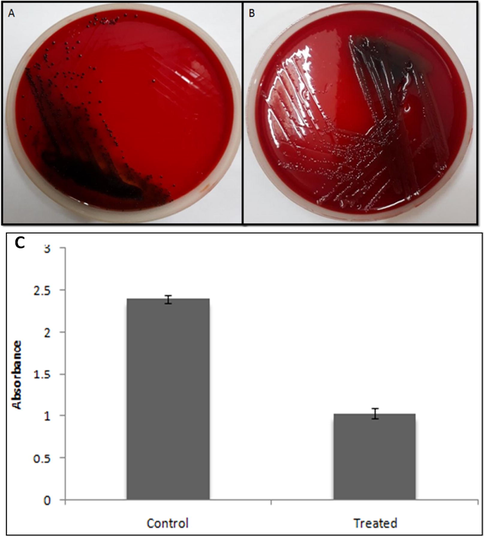
Effect of ZnO nanoparticles on biofilm of P. aeruginosa PAO1 using CRA and crystal violet method. (A) The appearance of black colonies representing exopolysaccharide production i.e. biofilm formation; (B) Treatment with ZnO NPs (300 µg/mL) showing the loss of black consistency i.e. inhibition of biofilm. (C) Showing quantitative assessment of effects of ZnO NPs on biofilm inhibition at 300 µg/mL.
![CLSM showing antibiofilm activity of ZnO NPS against PAO1. [A] Control; [B] treated with 300 µg/mL of ZnO NPs.](/content/184/2021/14/4/img/10.1016_j.arabjc.2021.103044-fig6.png)
CLSM showing antibiofilm activity of ZnO NPS against PAO1. [A] Control; [B] treated with 300 µg/mL of ZnO NPs.
Different components of the biofilm matrix-like exopolysaccharide and glucan are closely associated with the biofilm matrix and the virulence of bacteria (Rainey et al., 2019). Thus the reduction in the motility of bacterium results in the decrease in the formation of biofilm. Our results are indicative of a decrease in motility as well as a decrease in EPS production which is a prerequisite condition for the formation of biofilm. Our results are also in accordance with the previous studies where ZnO NPs effectively reduced motility function and biofilm formation in P. aeruginosa (Al-Shabib et al., 2018). Previous studies of Alavi et al. (Alavi et al., 2019) have also shown that green synthesized ZnO NPs effectively reduced biofilm formation in P. aeruginosa. García-Lara et al. (2015) have also shown that ZnO NPs effectively reduced the QS based virulence in P. aeruginosa. Ali et al. have also shown that green synthesized ZnO NPs effectively reduced the QS mediated virulence viz. pyocyanin, protease and hemolysin (Ali et al., 2020).
3.4 In silico analysis
Further to analyze the binding site of zinc oxide nanoparticles molecular docking analysis was also performed. In P. aeruginosa the Las and Rhl system along with their auto-inducers 3-oxo-C12-HSL and C4-HSL respectively regulates the QS mechanism. In the Las mechanism LasI regulates the production of 3-oxo-C12-HSL which binds to its receptor LasR and further governs virulence attributes like LasB elastase, protease, etc. In the Rhl system, RhlI regulates the synthesis of C4-HSL which further regulates the expression of lasA, lasB, pyocyanin, rhamnolipid (Chatterjee et al., 2016). Since most of the virulence of P. aeruginosa is governed by Las/Rhl system therefore these systems were considered as a molecular target for ZnO nanoparticles. To better understand the mechanism involved molecular docking analysis was performed to predict the possible sites of ZnO binding with AHL synthase and LasR and RhlR receptors.
The 3D structure was retrieved from Protein Data Bank for LasI (PDB ID: 1RO5; Fig. 7a) synthase and its transcriptional activator LasR (PDB ID: 2UV0; Fig. 8a). The 3D structure of RhlI/R synthase were not available in protein databank therefore RhlI synthase (Fig. 9a) and RhlR (Fig. 10a) were modeled using SWISS-MODEL homology. The validation of RhlI Synthase and RhlR model was analyzed by Ramachandran Plot; RhlI Synthase showed 94.5% of the residues in the favored region (Fig. 9b), similarly, RhlR showed 97.2% of the residues in the favored region (Fig. 10b).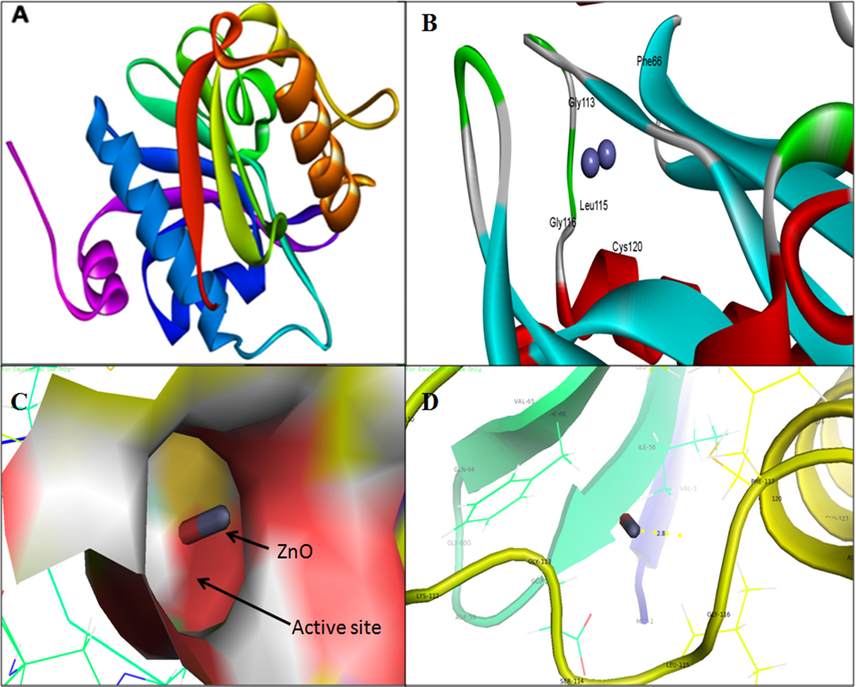
3D structure of Acyl-homoserine lactone (AHL) Synthase LasI (A); binding of ZnO NPs with the active site of LasI (B); close view of catalytic site of LasI, with bound ZnO NPs represented in capsular shape (C) Amino acids residues (Gly-116) of LasI involved in the interaction with ZnO NPs (D); LasI active site has been depicted by PyMol viewer. Interaction analysis of ZnO NPs with LasI has been explored through PatchDock.
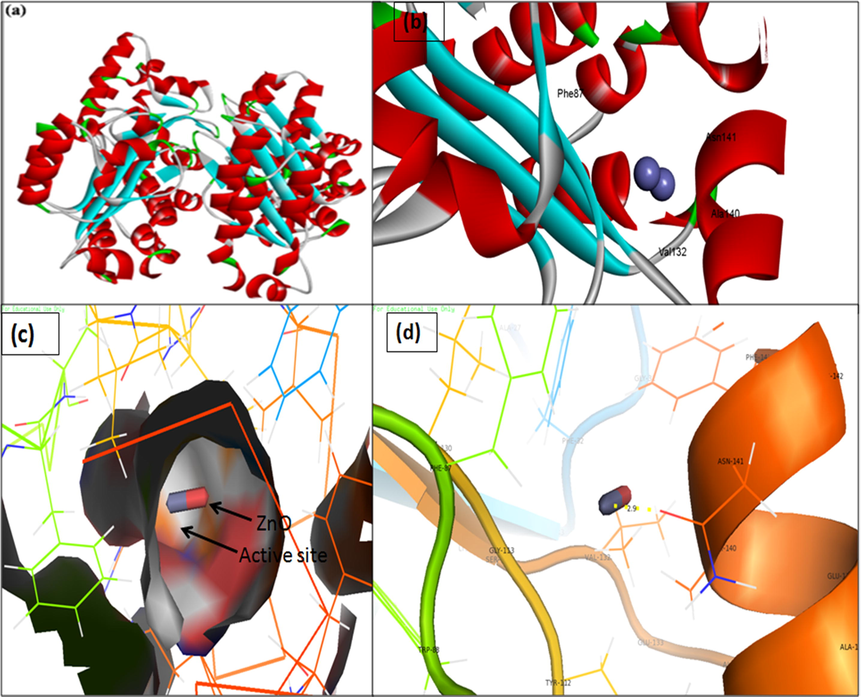
3D structure of transcriptional activator protein LasR. (a) binding of ZnO with the active site of LasR; (b) close view of catalytic site of LasR, with bound ZnO NPs represented as capsular structure; (c) Amino acids residues (Asn-141) of LasR involved in the interaction with ZnO NPs; (d) LasR active site has been depicted by PyMol viewer. Interaction analysis of ZnO NPs with LasR has been explored through PatchDock.
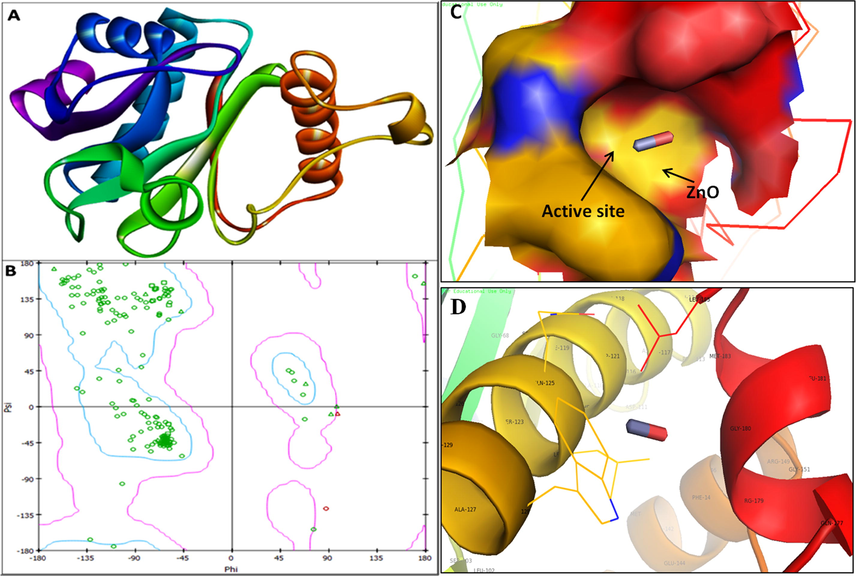
3D structure of transcriptional activator protein RhlI (A); RAMPAGE validation of conformation of RhlI (B); binding of ZnO with the active site of RhlI and close view of catalytic site of RhlI, with bound ZnO NPs represented as capsular structure (C). Amino acids residues (Gly-180) of RhlI involved in the interaction with ZnO NPs (D). RhlI active site has been depicted by PyMol viewer. Interaction analysis of ZnO NPs with RhlI has been explored through PatchDock.
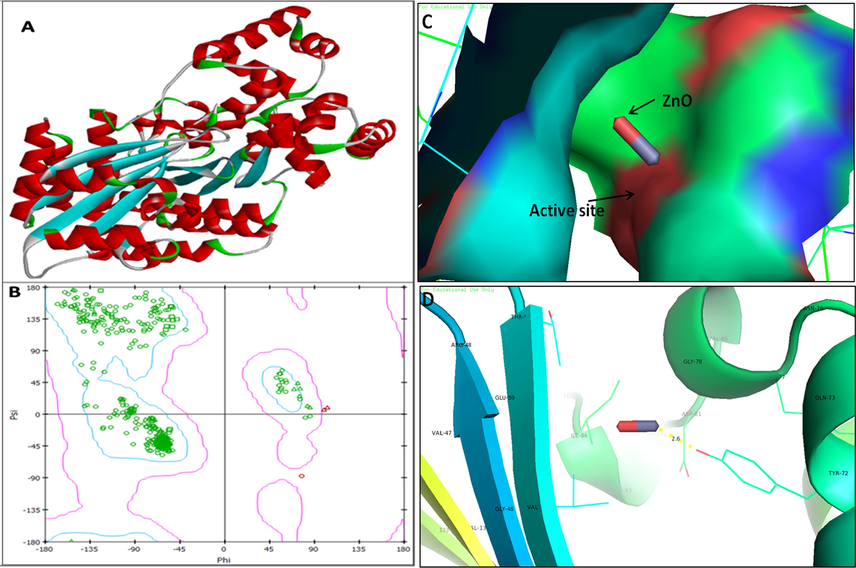
3D structure of regulatory protein RhlR (A); RAMPAGE validation of conformation of RhlR (B); binding of ZnO NPs with the active site of RhlR and close view of catalytic site of RhlR, with bound ZnO NPs represented as capsular structure (C). Specific residue (Tyr-72) of RhlR involved in the interaction with ZnO NPs (D); RhlR active site has been depicted by PyMol and the interaction analysis of ZnO NPs with RhlR has been explored by PatchDock.
The molecular analysis showed that ZnO was docked into the AHLs synthase LasI (Fig. 7b) and RhlI (Fig. 9c) and also to their regulatory activator proteins LasR (Fig. 8b) and RhlR (Fig. 10c). The molecular docking analysis showed that the ZnO binds to the catalytic Glycine residue of synthase LasI (Gly-116-ZnO = 2.8 Å), Asparagine residue of LasR receptor (Asn 141-ZnO = 2.9 Å), Glycine residue of RhlI synthase (Gly180-ZnO = 3.7 Å) and Tyrosine residue of RhlR receptor (Tyr72-ZnO = 2.6 Å). The atomic contact energy (ACE in Kcal/mol) and patch dock score obtained during ZnO NPs interaction with AHLs synthase i.e., LasI and RhlI and their regulatory activator proteins LasR and RhlR analyzed by PatchDock server was shown in Table 1.
S. No
Complexes (receptor + ZnO NPs)
ACE (Kcal/mol)
PatchDock Score
1.
LasI (PDB ID: 1RO5)
−9.16
754
2.
LasR (PDB ID: 2UV0)
−9.58
654
3.
RhlI synthase
−6.02
546
4.
RhlR
−6.79
550
Our analysis of molecular docking in P. aeruginosa reveals that ZnO binds to the catalytic cleft of AHL synthase and its receptors LasR/RhlR leading to the inhibition of QS controlled virulence factors. Since this is the first in silico report of ZnO interaction with QS therefore it is not possible to correlate it with previous findings. Although our results are consistent with the previous findings of Vyshnava et al. (Vyshnava et al., 2016) where they showed that silver got bound with Las R and RhlR and suppressed virulence and biofilm formation. Similar findings by Ali et al. (Ali et al., 2017) have been reported regarding the silver (Ag0) interaction with the QS system. They concluded that Ag got bound to the LasI and RhlI synthase and also with LasR and RhlR thereby inhibited the QS mechanism and finally the suppression of virulence.
4 Conclusions
The green synthesized ZnO NPs effectively inhibited the growth and QS mediated virulence in P. aeruginosa such as swarming, swimming and biofilm formation. The computational analysis (molecular docking) showed that ZnO NPs interrupted QS mechanism in P. aeruginosa by blocking the AHLs production that impede the synthesis of AHL Synthase i.e., LasI/RhlI and regulatory receptor proteins LasR/RhlR. The molecular docking studies revealed that green synthesized ZnO NPs effectively interacted with QS (Las/Rhl) mechanism and got bound to the catalytic cleft of LasI synthase (Gly-116-ZnO = 2.8 Å), RhlI synthase (Gly180-ZnO = 3.7 Å) and transcription receptor protein LasR receptor (Asn 141-ZnO = 2.9 Å), RhlR receptor (Tyr72-ZnO = 2.6 Å). Thus interaction of ZnO NPs with Las and Rhl system led to the inactivation of autoinducers (AHL) molecules which finally led to the inactivation of QS and down regulation of virulence factors. We are of opinion that ZnO NPs could be effectively used as alternative antimicrobial agents to prevent the colonization, biofilm formation and QS-mediated virulence by coating on medical devices, catheters and other medical- devices associated infections caused by pathogenic drug resistant bacterial and fungal species.
Acknowledgments
The authors would like to acknowledge Viral Research Diagnostic Laboratoty (DHR-ICMR) in Aligarh Muslim University, Aligarh, U.P. India. The authors also extend their appreciation to the Deputyship for Research & Innovation, “Ministry of Education” in Saudi Arabia for funding this research work through the project number IFKSURG-032.The authors would also like to acknowledge IRMC, Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia for providing support in conducting SEM and TEM analysis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Biogenic synthesis of metallic nanoparticles and prospects toward green chemistry. Dalton Trans.. 2015;44:9709-9717.
- [Google Scholar]
- Al-Shabib, N.A., Husain, F.M., Hassan, I., Khan, M.S., Ahmed, F., Qais, F.A., Oves, M., Rahman, M., Khan, R.A., Khan, A., 2018. Biofabrication of zinc oxide nanoparticle from Ochradenus baccatus leaves: broad-spectrum antibiofilm activity, protein binding studies, and in vivo toxicity and stress studies. J. Nanomater. 2018.
- Antibacterial, antibiofilm, antiquorum sensing, antimotility, and antioxidant activities of green fabricated Ag, Cu, TiO2, ZnO, and Fe3O4 NPs via protoparmeliopsis muralis lichen aqueous extract against multi-drug-resistant bacteria. ACS Biomater. Sci. Eng.. 2019;5:4228-4243.
- [Google Scholar]
- Effect of Biosynthesized ZnO Nanoparticles on Multi-Drug Resistant Pseudomonas Aeruginosa. Antibiotics. 2020;9:260.
- [Google Scholar]
- Antiquorum sensing activity of silver nanoparticles in P. aeruginosa: an in silico study. In Silico Pharmacology. 2017;5:12.
- [Google Scholar]
- Antiquorum sensing and antimicrobial activity of natural agents with potential use in food. J. Food Saf.. 2012;32:379-387.
- [Google Scholar]
- Biosynthesized ZnO-NPs from Morus indica attenuates methylglyoxal-induced protein glycation and RBC damage: In-vitro, in-vivo and molecular docking study. Biomolecules. 2019;9:882.
- [Google Scholar]
- Gum arabic capped-silver nanoparticles inhibit biofilm formation by multi-drug resistant strains of Pseudomonas aeruginosa. J. Basic Microbiol.. 2014;54(7):688-699.
- [Google Scholar]
- Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343-350.
- [Google Scholar]
- Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. Int. J. Med. Microbiol.. 2016;306:48-58.
- [Google Scholar]
- Variations in motility and biofilm formation of Salmonella enterica serovar Typhi. Gut Pathogens. 2014;6:2.
- [Google Scholar]
- Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol.. 2001;67:1865-1873.
- [Google Scholar]
- Inhibition of quorum-sensing-dependent virulence factors and biofilm formation of clinical and environmental P seudomonas aeruginosa strains by ZnO nanoparticles. Lett. Appl. Microbiol.. 2015;61:299-305.
- [Google Scholar]
- ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiol. Res.. 2014;169:888-896.
- [Google Scholar]
- Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis. 2009;30:S162-S173.
- [Google Scholar]
- Hutchison, M.L., Govan, J.R., 1999. Pathogenicity of microbes associated with cystic fibrosis. Microbes and infection1, 1005-1014.
- Apoptosis inducing ability of silver decorated highly reduced graphene oxide nanocomposites in A549 lung cancer. Int. J. Nanomed.. 2016;11:873.
- [Google Scholar]
- Antibacterial properties of silver nanoparticles synthesized using Pulicaria glutinosa plant extract as a green bioreductant. Int. J. Nanomed.. 2014;9:3551.
- [Google Scholar]
- Anti-quorum sensing and anti-biofilm activity of zinc oxide nanospikes. ACS Omega. 2020;5(50):32203-32215.
- [Google Scholar]
- Plant extracts as green reductants for the synthesis of silver nanoparticles: lessons from chemical synthesis. Dalton Trans.. 2018;47:11988-12010.
- [Google Scholar]
- The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors. Microbiology. 2005;151:3589-3602.
- [Google Scholar]
- Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol.. 2000;182:5990-5996.
- [Google Scholar]
- Biofabrication of zinc oxide nanoparticles from Melia azedarach and its potential in controlling soybean seed-borne phytopathogenic fungi. Saudi J. Biol. Sci.. 2020;27:1923-1930.
- [Google Scholar]
- The Pseudomonas quinolone signal (PQS): not just for quorum sensing anymore. Front. Cell. Infect. Microbiol.. 2018;8:230.
- [Google Scholar]
- A new transcriptional repressor of the Pseudomonas aeruginosa quorum sensing receptor gene lasR. PLoS ONE. 2013;8:e69554
- [Google Scholar]
- An investigation into the effects of silver nanoparticles on antibiotic resistance of naturally occurring bacteria in an estuarine sediment. Marine Environ. Res.. 2009;68:278-283.
- [Google Scholar]
- Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol.. 1998;28:449-461.
- [Google Scholar]
- Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol.. 2005;187:5267-5277.
- [Google Scholar]
- Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol.. 1997;179:5756-5767.
- [Google Scholar]
- Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci.. 1999;96:11229-11234.
- [Google Scholar]
- UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem.. 2004;25:1605-1612.
- [Google Scholar]
- Tumoricidal and bactericidal properties of ZnONPs synthesized using cassia auriculata leaf extract. Biomolecules. 2020;10:982.
- [Google Scholar]
- Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol.. 1998;30:285-293.
- [Google Scholar]
- Structure validation by Calpha geometry: Phi, psi and Cbeta deviation. Proteins. 2003;50:437-450.
- [Google Scholar]
- Rainey, K., Michalek, S.M., Wen, Z.T., Wu, H., 2019. Glycosyltransferase-mediated biofilm matrix dynamics and virulence of Streptococcus mutans. Appl. Environ. Microbiol. 85.
- Reuter, K., Steinbach, A., Helms, V., 2016. Interfering with bacterial quorum sensing. Perspect. Med. Chem. 8, PMC. S13209.
- Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol.. 1993;234:779-815.
- [Google Scholar]
- PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res.. 2005;33:W363-W367.
- [Google Scholar]
- Mycosynthesis of ZnO nanoparticles using Trichoderma spp. isolated from rhizosphere soils and its synergistic antibacterial effect against Xanthomonas oryzae pv. oryzae. J. Fungi. 2020;6(3):181.
- [Google Scholar]
- Mycogenic synthesis of extracellular zinc oxide nanoparticles from xylaria acuta and its nanoantibiotic potential. Int. J. Nanomed.. 2020;15:8519.
- [Google Scholar]
- Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis.. 1998;4:551.
- [Google Scholar]
- Effect of silver nanoparticles against the formation of biofilm by Pseudomonas aeruginosa an in silico approach. Appl. Biochem. Biotechnol.. 2016;180:426-437.
- [Google Scholar]
- 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res.. 2010;38:W469-W473.
- [Google Scholar]







