Cellulose filter paper immobilized α-glucosidase as a target enzyme-oriented fishing tool for screening inhibitors from Cyclocarya paliurus leaves
⁎Corresponding authors. 1282664933@qq.com (Zhao-Hui Guo), chenjuan@lzu.edu.cn (Juan Chen)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Cyclocarya paliurus leaves have exhibited a good hypoglycemic activity, however, the active compounds and inhibitory mechanisms against α-glucosidase remained unclear. In this study, a target enzyme-oriented fishing strategy based on cellulose filter paper (CFP) immobilized α-glucosidase combined with ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS/MS) and molecular docking was developed to screen and identify potential α-glucosidase inhibitors from C. paliurus leaves. Cellulose filter paper (CFP) as the carrier was modified by polydopamine/polyethyleneimine (PDA/PEI) co-deposition to form a uniform positive charge coating on the surface, and then α-glucosidase was immobilized on the modified CFP by electrostatic adsorption. By virtue of its instantaneous separation characteristic, the CPF-immobilized α-glucosidase was used as a target enzyme-oriented fishing tool to rapidly capture active compounds bound to α-glucosidase from the complex plant system. 36 active compounds were fished out from 70% ethanol fraction of C. paliurus leaves (IC50, 17.81 µg/mL), and further characterized by UPLC-QTOF-MS/MS. Furthermore, molecular docking was employed to predict inhibitory mechanisms, and the result showed that cyclocarioside A, cypaliuruside K, cypaliuruside J, cyclocarioside C, cyclocarioside I and pterocaryoside A could effectively interact with α-glucosidase by forming hydrogen bonds, hydrophobic bindings and Van der Waals force, and affinity energies ranged from −9.4 to −8.0 kJ/mol. Such a target enzyme-oriented fishing strategy would open a pathway for discovering targeted active compounds from medicinal plants.
Keywords
Target enzyme-oriented fishing tool
α-Glucosidase inhibitors
Cyclocarya paliurus
UPLC-QTOF-MS/MS
Molecular docking
1 Introduction
Diabetes mellitus (DM) is a common metabolic disorder, which has become an increasingly serious problem worldwide. DM is a chronic and progressive disease with high glucose levels in the blood and may lead to various complications such as heart attack, stroke, kidney failure, vision loss, and nerve damage (Abdullah et al., 2020; Liu et al., 2021a). Type-2 DM, as the most prevalent form of diabetes, is rising rapidly worldwide, and its main pathological symptom is abnormal regulation of postprandial blood glucose (Liu et al., 2021a). It is very necessary for alleviating the occurrence of Type-2 DM to control properly postprandial hyperglycemia by reducing the digestion and absorption of carbohydrates. α-Glucosidase is an intestinal enzyme that plays an important role in the carbohydrate metabolism by decomposing disaccharides and oligosaccharides into monosaccharides, increasing the postprandial blood sugar levels (Hu et al., 2021; Wang et al., 2020). Recently, α-glucosidase inhibitors have been utilized as clinical drugs to inhibit α-glucosidase activity and then control postprandial hyperglycemia associated with Type-2 DM. Although several synthetic α-glucosidase inhibitors such as acarbose, voglibose, and miglitol have good hypoglycemic effects, they bring a variety of adverse effects such as distension, diarrhea, and abdominal pain (Balfour et al., 1993; Liu et al., 2021b). Natural α-glucosidase inhibitors have become a hot research topic in the fields of medicine, pharmacy and functional food due to some advantages of safety, high efficacy, low side-effect and low cost (Cai et al, 2020; Liu et al., 2021b; Shen et al., 2020; Wang et al., 2020).
Cyclocarya paliurus, as a traditional edible and medicine plant, belongs to the Juglandaceae family originated in China, and has been commonly known as sweet tea tree. Leaves of C. paliurus have been consumed as a nutraceutical tea in China, and it is the first health tea from China that has been approved by the US Food and Drug Administration (FDA) (Wang et al., 2012). C. paliurus also was authorized as a new food raw material by National Health and Family Planning Commission of China in 2013 (Xie et al., 2016). Additionally, C. paliurus are known as “The Third Tree” and “The Giant Panda” in the medical field as traditional Chinese medicine (TCM) for the treatment of DM, inflammatory diseases, hyperlipidemia and hypertension (Kurihara et al., 2003; Li et al., 2021a; Zhao et al., 2019; Mahmood et al., 2013) due to a lot of active components, such as triterpenoids (Li et al., 2021a), polysaccharides (Yao et al., 2020), flavonoids (Ning et al., 2019) and phenolics (Zhang et al., 2010). Some studies have reported that the extract of C. paliurus and its active compounds had the good hypoglycemic effect on Type-2 DM mice, and could significantly inhibit α-glucosidase activity (Kurihara et al., 2003; Li et al., 2021a; Sun et al., 2020; Zhao et al., 2019; Chen et al., 2023). However, although C. paliurus leaves may be a promising source for natural α-glucosidase inhibitors, the complexity of extracts needs analytical technologies to screen and identify the bioactive compounds binding to α-glucosidase. Therefore, rapid and effective screening approaches for natural enzyme inhibitors from TCMs are still to be developed.
The affinity-based screening approach is regarded as an advanced, specific and high-efficiency technique for screening potential enzyme inhibitors from complex mixtures, compared with the conventional strategy with multistage separation. The strategy is based on affinity interactions between potential inhibitors and corresponding enzymes for the target enzyme-oriented drug discovery from complex mixtures (Hou et al., 2020), such as ultrafiltration (Cai et al., 2020; Ning et al., 2019), hollow fiber adsorption (Chen et al., 2017), and ligand fishing technology (Li et al., 2021b; Wan et al., 2021; Wang et al., 2018b). Recently, ligand fishing technology has played a significant role in capturing potential inhibitors from TCMs (Li et al., 2021b; Shen et al., 2020; Wan et al., 2021; Zhao et al., 2020). For example, some previous studies have developed the ligand fishing methods based on magnetic nanoparticles immobilized α-glucosidase to successfully screen natural α-glucosidase inhibitors from TCMs (Shen et al., 2020; Wang et al 2018b; Wubshet et al., 2019). However, although magnetic materials have been widely utilized as enzyme immobilization carriers, the major limitation is to be separated from the reaction system by adding an external magnetic field, resulting in the process complexity (Irfan et al., 2017; Mahmood et al., 2014, 2018; Shen et al., 2020; Wan et al., 2021). In our group, cellulose filter paper (CFP) has been used as an immobilization carrier to successfully fish out acetylcholinesterase and α-glucosidase inhibitors from TCMs by virtue of some good advantages of CFP-immobilized enzyme, especially, the instantaneously-separated characteristics, thus making the carrier preparation, enzyme immobilization and separation process be greatly simplified, compared with the magnetic nanoparticle carriers (Li et al., 2021b; Zhao et al., 2020; Liu et al., 2019).
In this study, a ligand fishing method was developed to specially capture active components against α-glucosidase from C. paliurus leaves based on CFP-immobilized α-glucosidase as a target enzyme-oriented fishing tool, combined with ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS/MS) and molecular docking. CFP carrier was modified by polydopamine/polyethyleneimine (PDA/PEI) co-deposition method to form a uniform positive charge coating on the surface, and then α-glucosidase was immobilized on the modified CFP by electrostatic adsorption (Li et al., 2021b). Firstly, the screening feasibility of CFP-immobilized α-glucosidase was verified by an artificial model mixture including α-glucosidase inhibitors and non-inhibitors. Secondly, CFP-immobilized α-glucosidase was incubated with the extract of C. paliurus leaves to fish out active compounds binding to α-glucosidase, which were further analyzed and identified by UPLC-QTOF-MS/MS. Moreover, inhibitory activities of screened active compounds on α-glucosidase were evaluated by the enzyme inhibition assay in vitro. The possible inhibitory mechanisms of potential inhibitors and α-glucosidase were further illuminated by molecular docking. The schematic illustration of the developed strategy is shown in Fig. 1.
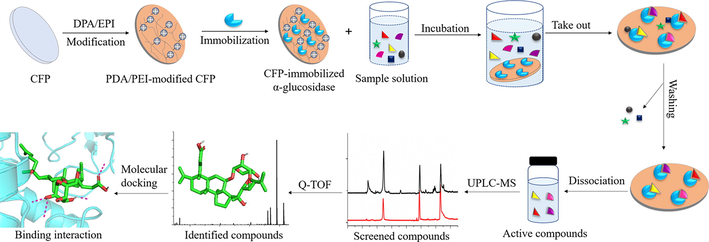
- Screening and identification of α-glucosidase inhibitors from Cyclocarya paliurus leaves using immobilized enzyme on cellulose filter papers, UPLC-QTOF-MS/MS and molecular docking.
2 Materials and methods
2.1 Materials
α-Glucosidase (EC 3.2.1.20, G8823-1KU) was extracted from yeast and obtained from Solarbio Technology Co., Ltd. (Shanghai, China). p-Nitrophenyl-α-D-glucopyranoside (p-NPG) was supplied by YuanYe Biotechnology Co., Ltd. (Shanghai, China). Standards of (+)-catechin, ferulic acid and quercetin were purchased by National Institutes for Food and Drug Control (Beijing, China), and kaempferitrin and arjunolic acid were obtained from the Chengdu Herbpurify Co. Ltd. (Chengdu, China). Dopamine hydrochloride (PDA) and polyethyleneimine (PEI) were supplied by Macklin Biochemical Co., Ltd. (Shanghai, China). Tris (hydroxy-methyl) aminomethane (Tris, 99%) were obtained from Solarbio Technology Co., Ltd. (Shanghai, China). The medium-speed qualitative filter paper was purchased from Fushun Civil Affairs Filter Paper Factory (Liaoning, China). D101 macroporous resin was purchased from an industrial chemical company affiliated to Nan Kai University (Tianjin, China). Acetonitrile and formic acid were of HPLC grade and obtained from Millipore Co. (Merck, Darmstadt, Germany). Ultrapure water was prepared by a Milli‐Q water purification system (Millipore, Bedford, MA, USA).
2.2 Instruments and chromatographic method
The chromatographic analysis was carried out by an Agilent 1290 Ultra performance liquid chromatograph (UPLC) system coupled with quaternary solvent delivery system, auto-sampler, column oven and vacuum degasser. Chromatographic separation was performed on an Agilent ZORBAX Eclipse Plus C18 column (3.0 mm × 150 mm, 1.8 µm), and column temperature was set at 35℃. 0.1% formic acid in acetonitrile and 0.1% formic acid in water were selected as the mobile phase at a flow rate of 0.20 mL/min. For the analysis of the artificial model mixture, gradient elution program was as follows: 0–15 min, 20–80% A. For 70% eluent fraction of C. paliurus leaves, gradient elution program was as follows: 0–3 min, 20–40 % A; 3–5 min, 40–50 % A; 5–15 min, 50–75 % A; 15–45 min, 75–100 % A; 45–50 min, 100 % A (Li et al., 2022; He et al., 2022); Each solution was filtrated, and the volume of injection was 2 μL.
An Agilent 6545 quadrupole time-of-flight (QTOF) tandem mass spectrometer (Agilent Technologies, USA) equipped with a Dual AJS electrospray ionization source (ESI) was performed for MS analysis in negative ion mode. The acquisition parameters for MS/MS analysis were set as follows. Both the drying gas temperature and sheath gas temperature were maintained at 350 ℃. The flow rates of drying gas and sheath gas were 12.0 L/min and 10.0 L/min, respectively. The capillary voltage was set at 3.5 kV. The fragmentor voltage and skimmer voltage were 130 V and 65 V, respectively. The mass spectra were recorded at a scan range of m/z 100–1000, and the collision energies of MS2 spectra were optimized from 10 to 50 eV.
2.3 Extraction and purification of C. paliurus leaves
Dried C. paliurus leaves were purchased from Zhangjiajie, Anhui Province, in China and identified by Prof. Ling Jin (College of Pharmacy, Gansu University of Chinese Medicine, Lanzhou, China). The extraction method of C. paliurus leaves was according to previous studies (He et al., 2022; Sun et al., 2020). Samples (50 g) were extracted twice by ultrasonication with 60% ethanol (450 mL and 350 mL) for 60 min each time. The extraction solutions were combined, filtrated and then concentrated to dryness by a rotary vacuum evaporator under reduce pressure. The residue was re-dissolved in 5% ethanol solution with the final concentration of 50 mg/mL. The obtained sample solution was transferred to column chromatography (30 × 3 cm) on D101 macroporous resin, and then eluted successively using water and 30, 50, 70 and 95% ethanol solutions. Five fractions were condensed and stored in a refrigerator at 4℃.
2.4 Preparation of α-glucosidase immobilized on CFP
α-Glucosidase immobilization was performed according to the method of our group (Li et al., 2021b). Briefly, the CFP was immersed into PDA and PEI (2 mg/mL each) dissolved 0.1 M tris buffer (pH 8.5), and coated for 7 h at 150 r/min and 30 °C to obtain PDA/PEI-modified CFP. Then, the modified CFPs (Diameter, 6 mm) were immersed into 0.05 mg/mL of α-glucosidase and shaken for 1 h at 150 r/min and 30 °C. The obtained CFP-immobilized α-glucosidase were washed several times with PBS (pH 6.0) and dried at 35 °C for subsequent screening.
2.5 Ligand fishing evaluation by an artificial model mixture
Artificial model compounds, which consisted of (+)-catechin (1) (Fu et al., 2021), ferulic acid (2) (Wang et al., 2018b) and quercetin (3) (Shen et al., 2020), were dissolved in 20 mM PBS (pH 8.0) containing 5 % DMSO to obtain the model solution (S0, 1 mg/mL each). 6 discs of CFP-immobilized α-glucosidase were incubated with 2 mL of the model solution for 30 min at 70 °C. After incubation, the CFP discs were directly taken out by a tweezer and washed with 3 × 600 μL of PBS buffer to obtain the washing solutions (S1-S3). Then, the discs were eluted with 3 × 600 μL of 80% methanol for 10 min each time to obtain dissociation solutions (S4-S6). Solutions (S0, S1-S3 and S4-S6) were analyzed by UPLC-QTOF-MS/MS.
2.6 Screening α-glucosidase inhibitors from C. paliurus leaves by ligand fishing
Both 70% eluent fraction of C. paliurus leaves and quercetin as the positive control were dissolved in PBS (20 mM, pH 8.0) containing 5 % DMSO to obtain the mixed solution with final concentrations of 2.0 mg/mL and 1 mg/mL, respectively. 2 mL of the mixed solution was incubated with 6 discs of CFP-immobilized α-glucosidase for 30 min at 70 °C. Active compounds, which exhibited specific affinity interactions with α-glucosidase, would form enzyme-inhibitor complexes, while inactive components with no or low affinity to α-glucosidase would keep free states. After incubation, the CFPs discs were instantaneously taken out and then washed three times with 600 μL of PBS to remove inactive components. Afterwards, the CFPs discs were immersed in 600 μL of 80% methanol for three times, 10 min each time, and then active compounds were dissociated from complexes. The dissociation solutions were collected and evaporated, and then the residue was re-dissolved in 80% methanol for UPLC-QTOF-MS/MS analysis.
2.7 α-Glucosidase inhibition assay
The inhibition activity of potential inhibitors on α-glucosidase was evaluated according to the method with some modification (Shen et al., 2020). α-Glucosidase and p-NPG were dissolved in 0.1 M PBS (pH 6.86) to obtain concentrations of 1 U/mL and 5.0 mM, respectively. 40 µL of α-glucosidase, 40 µL of potential inhibitors and 80 µL of PBS were pre-incubated for 10 min at 37 ℃. Then, 40 µL of p-NPG was added into the mixture to initiate the enzymatic reaction, and the mixture was incubated for 20 min. The absorbance of the mixture was measured at 405 nm. The inhibition rate (%) was calculated according to equation:
Where Ac, Abc, Ai and Abi present absorbances of the mixture (enzyme without inhibitor, no enzyme and inhibitor, enzyme and inhibitor, and inhibitor without enzyme), respectively.
2.8 Molecular docking
Molecular docking was carried out to further investigate affinity interactions between potential inhibitors and α-glucosidase by Autodock 4.2 software. According to the previous studies (Ning et al., 2019; Shen et al., 2020), α-glucosidase (PDB ID: 3A4A) was utilized to perform the docking assay, and its crystal structure was obtained from the Protein Data Bank ( https://www.rcsb.org/pdb). The original ligands and water of α-glucosidase were removed by PyMOL software. The hydrogen atoms and gasteiger charges were added to α-glucosidase, and then the file was saved as receptor.pdbqt by AutoDockTools. The 3D structures of potential inhibitors were acquired by ChemBio3D Ultra 14.0, and then their torsions along with rotatable bonds were assigned and the files were saved as ligand.pdbqt by AutoDockTools, respectively. The semi-flexible docking was performed by Autodock Vina (Hu et al., 2021; Li et al., 2022). The grid parameters were shown as follows: x = 25.38, y = -2.75 Å, z = 18.186, and the dimensions 52 × 68 × 62 Å (Hu et al., 2021). The docking result with the lowest binding energy and the highest scoring was selected to be the most optimal docking model of the α-glucosidase-inhibitor complex. The binding interactions between potential inhibitors and α-glucosidase were visualized using Discovery Studio 4.5 and PyMOL software.
3 Results and discussion
3.1 Ligand fishing evaluation by an artificial model mixture
The artificial model mixture, including (+)-catechin (1), ferulic acid (2) and quercetin (3), was used to investigate the feasibility of ligand fishing assay. Quercetin and (+)-Catechin as two known α-glucosidase inhibitors were employed as the positive controls (Wan et al., 2021; Shen et al., 2020; Fu et al., 2021), while ferulic acid as the negative control had a weak affinity to α-glucosidase (Wang et al., 2018b). The 6 discs of CFP-immobilized α-glucosidase were incubated with 600 μL of model solution (S0) to form enzyme-inhibitor complexes for 30 min at 70℃. After incubation, the CFP discs were instantaneously taken out by a tweezer. The discs were washed three times with 600 μL of PBS buffer to remove weak affinity to α-glucosidase from enzyme-inhibitor complexes (S1-S3), and then immersed in 600 μL of 80% methanol to denature α-glucosidase for the dissociation of strong α-glucosidase inhibitors (S4-S6). As shown in Fig. 2, quercetin and (+)-catechin as the positive control were successfully fished out from S0 and could be determined in dissociation solutions S4-S6. Quercetin could have the stronger affinity than (+)-catechin due to the amount of quercetin was higher than (+)-catechin which were released by denaturing CFP-immobilized α-glucosidase in S4-S6. However, ferulic acid as the negative control was not detected in S4-S6 at all, because it was washed gradually in washing solution S1-S3 due to its weak affinity to α-glucosidase. The result was just as we expected and similar with the literatures (Wang et al., 2018b; Shen et al., 2020), indicating that the affinity screening strategy based on ligand fishing and UPLC-QTOF-MS/MS was effective and reliable to screen natural α-glucosidase inhibitors in the artificial model mixture.
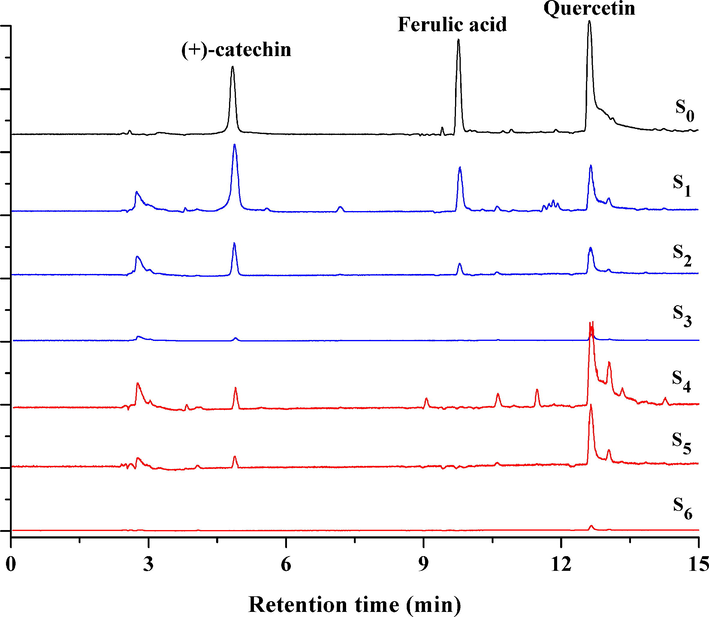
- Extract ion chromatograms of the artificial model mixture (S0), PBS washing solutions (S1-S3) and dissociation solutions (S4-S6) in the feasibility evaluation of ligand fishing assay.
3.2 Screening α-glucosidase inhibitors from C. paliurus leaves by ligand fishing assay
Enzyme inhibition assay in vitro was used to estimate inhibitory activities of five fractions of the crude extract from C. paliurus leaves on α-glucosidase. The results showed that the 70% ethanol fraction of C. paliurus leaves showed significant inhibitory activity against α-glucosidase, compared with the other four fractions. It was obvious that both 70% ethanol fraction and quercetin (as the positive control) inhibited α-glucosidase activity in a concentration-dependent manner and their IC50 values were calculated to be 17.81 ± 0.18 µg/mL and 4.15 ± 0.04 µg/mL, respectively. Results suggested that 70% ethanol fraction might contain active compounds against α-glucosidase.
The 70% ethanol fraction containing quercetin was incubated with CFP-immobilized α-glucosidase for 30 min at 70℃. Quercetin was used as the positive control to further verify reliability and practicality of the proposed ligand fishing method in the extract from C. paliurus leaves. After incubation, potential inhibitors would form α-glucosidase-inhibitor complexes, and then complexes were separated from the reaction solution in virtue of the instantaneously-separated characteristic of CFP. The discs were washed with PBS buffer (3 × 600 μL), and then active components would be released from the complexes by adding 80% methanol (3 × 600 μL). As shown in Fig. 3, a total of 37 peaks were determined in the chromatogram, implying that these compounds could have binding abilities to α-glucosidase (Fig. 2 B). In addition, quercetin was successfully captured and detected at 10.30 min (Fig. 2 C), suggesting that the ligand fishing assay is reliable and applicable for screening natural α-glucosidase from complex matrixes.
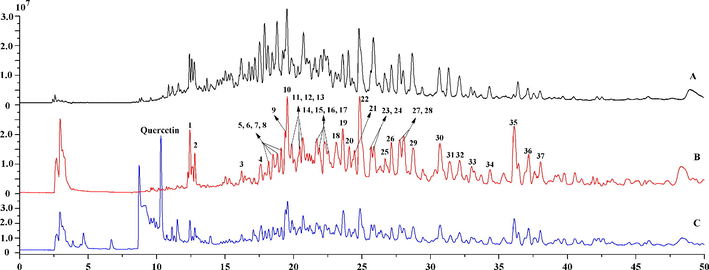
- Total ion chromatograms of 70% ethanol fraction of Cyclocarya paliurus leaves (A), screened active compounds by CFP-immobilized α-glucosidase (B), active components and quercetin (as positive control) fished out by CFP-immobilized α-glucosidase (C).
3.3 Identification of potential α-glucosidase inhibitors
Dammarane triterpenoids are characteristic indicators and active compounds of C. paliurus, and exhibited anti-diabetic, anti-hyperglycemia, anti-oxidative and anti-inflammatory effects (Li et al., 2021a; Sun et al., 2020; Zhou et al., 2021; Zhao et al., 2019). 3,4-seco-dammarane triterpenoids of C. paliurus are rare in natural plants, characterized by the breaking of the chemical bond between C-3 and C-4 of dammarane triterpenoids. The differences in the chemical structures of triterpenoids were mainly due to the existence of a variety of aglycones, and types of glycosyl moieties and their connecting positions on the aglycones. In the negative ion mode, dammarane triterpenoids, except for 3,4-seco-dammarane triterpenoids, often form addition ions [M + HCOO]− due to the addition of formic acid in the mobile phase. Retention time (Rt), calculated and measured molecular mass, adduct ion, mass error, MS/MS fragment ions, molecular formula, identification and references are summarized in Table 1. Chemical structures of potential α-glucosidase inhibitors are shown in Fig. 4.
| Peak no. | RT (min) | Calculated m/z [M−H]- | Measured m/z[M-H]- | Measured [M + HCOO]- | Error (ppm) | MS/MS (m/z) | Molecular formula | Identification | Reference |
|---|---|---|---|---|---|---|---|---|---|
| *1 | 12.44 | 577.1351 | 577.1332 | 3.37 | 431.0951[M-H-Rha], 285.0416[M-H-2Rha] | C27H30O14 | Kaempferitrin | Ning et al.,2019 | |
| 2 | 12.80 | 577.1351 | 577.1339 | 2.16 | 431.0967[M-H-Rha], 285.0402[M-H-2Rha] | C30H26O12 | Kaempferol 7,4′-dirhamnoside | ||
| *3 | 16.23 | 487.3429 | 487.3423 | 1.22 | 445.2943, 401.3056, 389.2689 | C30H48O5 | Arjunolic acid | ||
| 4 | 17.60 | 753.4795 | 753.4799 | 799.4943 | −0.59 | 621.4373[M-H-C5H8O4 at C3], 607.4206[M-H-C5H8O4-C6H10O4 at C12], 145.0519[Qui-H], 131.0339[Xyl/Ara Ara-H], 101.0231[Xyl/Ara-H2O-CH2O-H] | C41H70O12 | Cyclocarioside I/K/Z7 | Cui et al., 2015; Shu et al., 1995; Sun et al., 2020 |
| 5 | 18.57 | 795.4900 | 795.4913 | 841.4954 | −1.61 | 753.4765[M-H-C2H2O], 735.4661[M-H-C2H2O-H2O], 621.4350[M-H-C2H2O-C5H8O4 at C3], 607.4143[M-H-C2H2O-C6H10O4 at C12], 131.0349[Ara-H] | C43H72O13 | Cyclocarioside A/H Compound 2 Cypaliuruside V |
Li et al., 2012; Yan et al., 2021; Zhu et al., 2021 |
| 6 | 18.48 | 753.4795 | 753.4766 | 799.4842 | 3.78 | 621.4301[M-H-C5H8O4 at C3], 131.0345, 101.0237 | C41H70O12 | Cyclocarioside Z14 | Li et al., 2021a |
| 7 | 18.84 | 621.4008 | 621.3999 | 1.46 | 521.3105[M-H-C6H12O at C20], 489.3567[M-H-C5H8O4 at C12], 471.3475[M-H-C5H8O4-H2O], 389.2689[M-H-C5H8O4-H2O-C6H10], 371.2571[M-H-C5H8O4-2H2O-C6H10], 101.0229 | C35H58O9 | Pterocaryoside B Cyclocarioside J |
Cui et al., 2015; Kennelly et al., 1995 | |
| 8 | 19.25 | 621.4008 | 621.3995 | 2.1 | 521.3101, 489.3564, 471.3455, 389.2740, 371.2525, 101.0223 | C35H58O9 | Pterocaryoside B Cyclocarioside J |
||
| 9 | 19.39 | 753.4795 | 753.4799 | 799.4873 | −0.59 | 621.0.4354, 607.4183, 145.0517, 131.0350, 101.0243 | C41H70O12 | Cyclocarioside I/K/Z7 | |
| 10 | 19.52 | 753.4795 | 753.4791 | 799.4873 | 0.47 | 621.4358, 607.4192, 145.0519, 131.0348, 101.0244 | C41H70O12 | Cyclocarioside I/K/Z7 | |
| 11 | 19.87 | 721.4532 | 721.4536 | 767.4584 | −0.5 | 639.3748 [M−H−C6H10 at C20], 589.4122[M-H-C5H8O4 at C3], 507.3329[M-H-C5H8O4-C6H10], 131.0350[Ara-H], 101.0239 | C40H66O11 | Cyclocarioside Z5 | Sun et al., 2020 |
| 12 | 20.37 | 635.4165 | 635.4156 | 1.35 | 535.3264[M-H-C6H12O at C20], 489.3556[M-H-C6H10O4 at C12], 471.3481[M-H-C6H10O4-H2O], 389.2695[M-H-C6H10O4-H2O-C6H10], 371.2589[M-H-C6H10O4-2H2O-C6H10], 101.0234 | C36H60O9 | Pterocaryoside A | Kennelly et al., 1995 | |
| 13 | 20.68 | 753.4790 | 799.4854 | 621.4360[M-H-C5H8O4 at C3], 131.0334 [Ara-H], 101.0234 | C41H70O12 | Cyclocarioside Z16 | Li et al., 2021a | ||
| 14 | 21.98 | 663.4114 | 663.4103 | 1.61 | 621.3965[M-H-C2H2O], 603.3888[M-H-C2H2O-H2O], 489.3590[M-H-C2H2O-C5H8O4 at C24], 471.3457[M-H-C2H2O-C5H8O4], 131.0326[Ara-H], 101.0248 | C37H60O10 | Cyclocarioside O | Qin, 2018 | |
| 15 | 21.90 | 735.4672 | 735.4672 | 781.4722 | 2.29 | 653.3901[M-H-C6H10 at C20], 589.4088[M-H-C6H10O4 at C12], 507.3357[M-H-C6H10- C6H10O4], 101.0240 | C41H68O11 | Cyclocarioside P | Wang et al., 2018a |
| 16 | 22.24 | 781.4744 | 781.4733 | 827.4805 | 1.36 | 739.4614[M-H-C2H2O], 721.4523[M-H-C2H2O-H2O], 607.4223[M-H-C2H2O-C6H8O4 at C3], 131.0336, 101.0236 | C42H70O13 | Cyclocarioside C | Jiang et al., 2006 |
| 17 | 22.35 | 795.4900 | 795.4907 | 841.4958 | −0.86 | 753.4788, 735.4681, 621.4357, 607.4134, 131.0341 | C43H72O13 | Cyclocarioside A/H Compound 2 Cypaliuruside V |
|
| 18 | 23.08 | 633.4008 | 633.3997 | 1.74 | 551.3218[M-H-C6H10 at C20], 471.3454[M-H-C6H10O5 at C11], 453.3371[M-H-C6H10O5-H2O], 389.2680[M-H-C6H10-C6H10O5], 371.2582[M-H-C6H10-C6H10O5-H2O], 161.0453[Glu-H-H2O], 101.0236 | C36H58O9 | Cypaliuruside J | Zhou et al., 2021 | |
| 19 | 23.70 | 767.4951 | 767.4940 | 813.5012 | 1.43 | 621.4364[M-H-C6H10O4 at C3], 603.4203[M-H-C6H10O4-H2O], 145.0498[Qiu-H], 101.0239, 475.3769 | C42H72O12 | Cyclocarioside Z13 | Li et al., 2021a |
| 20 | 24.09 | 795.4900 | 795.4902 | 841.4953 | −0.23 | 753.4798, 735.4661, 621.4382, 607.4136, 101.0241 | C43H72O13 | Cyclocarioside A/H Compound 2 Cypaliuruside V |
|
| 21 | 24.46 | 621.4008 | 621.3994 | 2.26 | 489.3585[M-H-C5H8O4 at C12], 471.3475[M-H-C5H8O4-H2O], 131.0338, 101.0234 | C35H58O9 | Compound 1 | Fang et al., 2019 | |
| 22 | 24.90 | 795.4905 | 841.4941 | −0.62 | 753.4791, 735.4674, 621.4369, 607.4146, 131.0347 | C43H72O13 | Cyclocarioside A/H Compound 2 Cypaliuruside V |
||
| 23 | 25.65 | 621.4372 | 621.4352 | 667.4334 | 3.2 | 603.3354[M-H-H2O], 475.3350[M-H-C6H10O4 at C11], 101.02 | C36H62O8 | Cyclocarioside Z9 | Li et al., 2021a |
| 24 | 25.89 | 649.4321 | 649.4324 | −0.45 | 521.3118[M-H-2CH2-C6H12O], 517.3895[M-H-C5H8O4 at C11/12], 499.3787[M-H-C5H8O4-H2O], 389.2687[M-H-2CH2-C6H12O-C5H8O4], 371.2591[M-H-2CH2-C6H12O-C5H8O4-H2O], 131.0339, 101.0232 | C37H62O9 | Cypaliuruside D Cyclocarioside X |
Liu et al., 2020; Zhou et al., 2021 | |
| 25 | 26.71 | 603.3844 | 603.3896 | 1.06 | 521.3108[M-H-C6H10 at C20], 471.3480[M-H-C5H8O4 at C11/12], 453.3361[M-H-C5H8O4-H2O], 389.2685[M-H-C6H10-C5H8O4], 371.2570[M-H-C6H10-C5H8O4-H2O], 101.0233 | C35H56O8 | Cypaliuruside A Cyclocarioside I |
Cui et al., 2015; Zhou et al 2021 | |
| 26 | 27.16 | 795.4900 | 795.4917 | 841.4958 | −2.11 | 753.4783, 735.4667, 621.4378, 607.4135, 131.0342 | C43H72O13 | Cyclocarioside A/H Compound 2/ Cypaliuruside V |
|
| 27 | 27.75 | 603.3902 | 603.3890 | 2.06 | 521.3111, 471.3469, 453.3373, 389.2693, 371.2578, 101.0236 | C35H56O8 | Cypaliuruside A Cyclocarioside I |
||
| 28 | 28.04 | 809.5046 | 855.5112 | 1.32 | 767.4947[M-H-C2H2O], 749.4840[M-H-C2H2O-H2O], 621.4400[M-H-C2H2O-C6H10O4 at C11], 101.0246 | C44H74O13 | Cyclocarioside Z17 | Li et al., 2021a | |
| 29 | 28.72 | 663.4478 | 663.4474 | 0.54 | 535.3270[M-H-2CH2-C6H12O at C20], 517.3896[M-H-C6H10O4 at C11/12], 499.3787[M-H-C6H10O4-H2O], 389.2698[M-H-2CH2-C6H12O-C6H10O4], 371.2571[M-H-2CH2-C6H12O-C6H10O4-H2O], 101.0236 | C38H64O9 | Cypaliuruside K/S Cyclocarioside Y |
Liu et al., 2020; Zhou, et al., 2021; Zhu et al., 2021 | |
| 30 | 30.69 | 617.4059 | 617.4055 | 0.63 | 535.3266[M-H-C6H10 at C20], 471.3480[M-H-C6H10O4 at C12], 453.3385[M-H-C6H10O4-H2O], 389.2692[M-H-C6H10-C6H10O4], 371.2594[M-H-C6H10 -C6H10O4-H2O], 101.0245 | C36H58O8 | Cyclocarioside K | Wu et al., 2014 | |
| 31 | 31.35 | 809.5057 | 809.5036 | 855.5105 | 2.55 | 767.4936[M-H-C2H2O], 749.4832[M-H-C2H2O-H2O], 621.4257[M-H-C2H2O-C6H10O4 at C12], 101.0243 | C44H74O13 | Cyclocarioside N | Wu et al., 2017 |
| 32 | 32.05 | 663.4478 | 663.4483 | −0.82 | 535.3289, 517.3895, 499.3753, 389.2680, 371.2589, 101.0240 | C38H64O9 | Cypaliuruside K/S Cyclocarioside Y |
Zhou et al., 2021; Zhu et al., 2021; Liu et al., 2020 | |
| 33 | 33.00 | 645.4008 | 645.4004 | 0.63 | 603.3910[M-H-C2H2O], 585.3784[M-H-C2H2O-H2O], 453.3365[M-H-C2H2O-H2O-C5H8O4 at C20] | C37H58O9 | Cyclocarioside V | Qin, 2018 | |
| 34 | 34.35 | 649.4321 | 649.4315 | 0.93 | 521.3109, 517.3893, 499.3771, 389.2687, 371.2580, 101.0235 | C37H62O9 | Cypaliuruside D Cyclocarioside X |
Liu et al., 2020; Zhou et al., 2021 | |
| 35 | 36.13 | 339.2333 | 339.2321 | 2.51 | 163.1227 | C23H32O2 | unknown | ||
| 36 | 37.18 | 471.3480 | 471.3469 | 2.29 | 453.3361[M-H-H2O], 389.2690[M-H-2H2O-HCOOH] | C30H48O4 | Maslinic acid | ||
| 37 | 38.08 | 663.4475 | 663.4475 | 0.39 | 535.3271, 517.3898, 499.3791, 389.2698, 371.2555, 101.0234 | C38H64O9 | Cypaliuruside K/S Cyclocarioside Y |
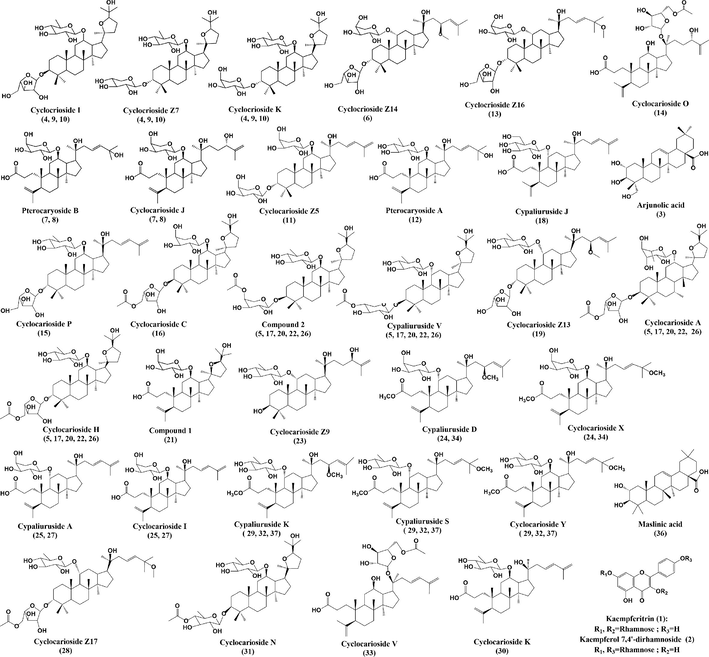
- Chemical structures of screened potential α-glucosidase inhibitors from Cyclocarya paliurus leaves.
Compound 5, 17, 20, 22 and 26 exhibited same adduct ions at m/z 841.49 [M + HCOO]− and molecular ions at m/z 795.49 [M−H]−, which produced fragment ions m/z 753.47 [M−H−42 Da]−, m/z 735.46 [M−H−42 Da-18 Da]−, 621.43 [M−H−42 Da-132 Da]−, m/z 607.41 [M−H−42 Da-146 Da]−, and m/z 131.03 [Ara-H2O-H]−, Hence, they were cyclocarioside A/H, compound 2 and cypaliuruside V isomers (Li et al., 2012; Yan et al., 2021; Zhu et al., 2021), and the MS/MS spectrum and fragmentation pattern of cyclocarioside H are shown in Fig. 5. Compound 16 had the adduct ion at m/z 827.47 [M + HCOO]− and molecular ion at m/z 781.47 [M−H]−. It gave fragment ions at m/z 739.46 [M−H−42 Da]−, m/z 721.45 [M−H−42 Da-18 Da]−, m/z 607.42 [M−H−42 Da-132 Da]−, m/z 131.03 [Ara-H2O-H]− and m/z 101.02 [Ara-H2O-CH2O-H]−, which was identified as cyclocarioside C (Jiang et al., 2006), and its MS/MS spectrum and fragmentation pattern are shown in Fig. S1.
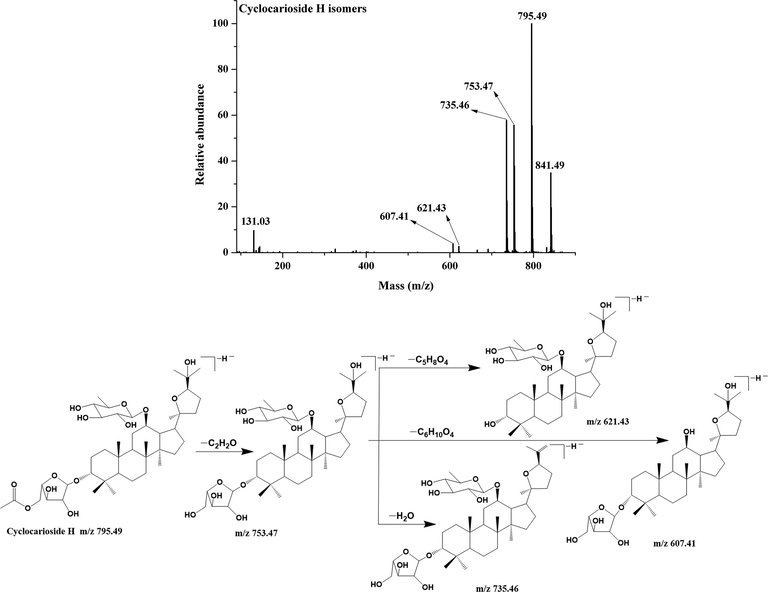
- MS/MS spectrum and fragmentation pattern of cyclocarioside H of C. paliurus leaves in negative ion mode.
Compound 12 gave the molecular ion at m/z 635.41 [M−H]−, which were identified as pterocaryoside A based on fragment ions at m/z 535.32 [M−H−100 Da]−, m/z 489.35 [M−H−146 Da]−, m/z 471.34 [M−H−146 Da-18 Da]−, m/z 389.26 [M−H−146 Da-18 Da-82 Da]−, m/z 371.25 [M−H−146 Da-18 Da-82 Da-18 Da]−, and m/z 101.02 (Kennelly et al., 1995). Its MS/MS spectrum and fragmentation pattern are shown in Fig.S2. Compound 29, 32 and 37 exhibited same molecular ions at m/z 663.44 [M−H]−, which were identified as cypaliuruside K/S and cyclocarioside Y isomers based on fragment ions at m/z 535.32 [M−H−28 Da-100 Da]−, m/z 517.38 [M−H−146 Da]−, m/z 499.37 [M−H−146 Da-18 Da]−, m/z 389.26 [M−H−28 Da-100 Da-146 Da]−, m/z 371.25 [M−H−28 Da-100 Da-146 Da-18 Da]− and m/z 101.02 (Zhou, et al., 2021; Liu et al., 2020; Zhu et al., 2021). The MS/MS spectrum and fragmentation pattern are shown in Fig. 6. Compound 25 and 27 exhibited same molecular ions at m/z 603.38 [M−H]−, and further produced fragment ions at m/z 521.31 [M−H−82 Da]−, m/z 471.34 [M−H−132 Da]−, m/z 453.33[M-H-132 Da-18 Da]−, m/z 389.26 [M−H−82 Da-132 Da]−, m/z 371.25 [M−H−82 Da-132 Da-18 Da]− and m/z 101.02. Hence, they are identified as cypaliuruside A and cyclocarioside I isomers (Cui et al., 2015; Zhou et al., 2021) and the MS/MS spectrum and fragmentation pattern are shown in Fig.S3. Compound 18 had molecular ion m/z 633.39 [M−H]−, which generated fragment ions at m/z 551.32 [M−H−82 Da]−, m/z 471.34 [M−H−162 Da]−, m/z 453.33 [M−H−162 Da-18 Da]−, m/z 389.26 [M−H−82 Da-162 Da]−, m/z 371.25 [M−H−82 Da-162 Da-18 Da]−, m/z 161.04 [Glu-H]−, and m/z 101.02. Therefore, it was identified as cypaliuruside J (Zhou et al., 2021), and the MS/MS spectrum and fragmentation pattern are shown in Fig. S4.
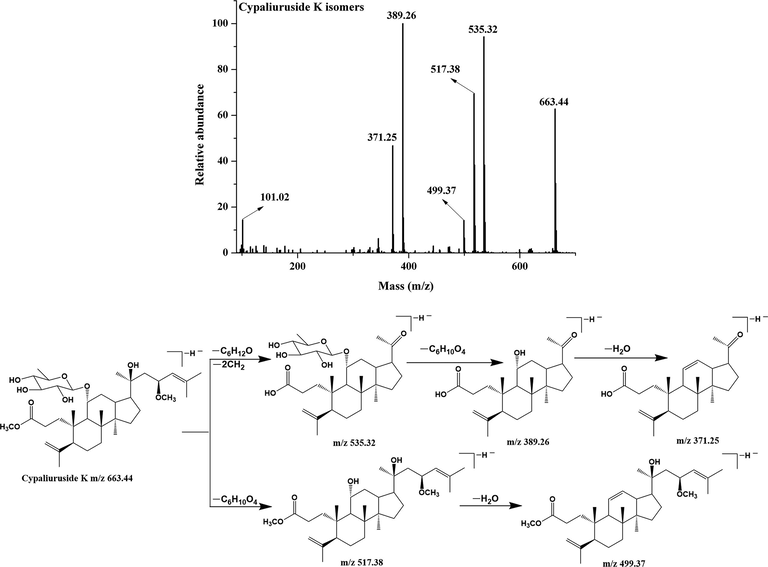
- MS/MS spectrum and fragmentation pattern of cypaliuruside K of C. paliurus leaves in negative ion mode.
3.4 α-Glucosidase inhibition assay in vitro
To further validate the reliability of ligand fishing assay, inhibitory activities of active compounds against α-glucosidase were evaluated by enzyme inhibition assay in vitro. However, it is worth mentioning that commercially available standards from C. paliurus are few. IC50 values of kaempferitrin and arjunolic acid were measured, and they are 1.12 mM and 339.02 μM, respectively. In addition to, cypaliuruside J, cypaliuruside D, cypaliuruside I, cypaliuruside K, cyclocarioside Z9, cyclocarioside Z13 and cyclocarioside Z14 showed remarkable inhibitory activities against α-glucosidase with IC50 values of 2.22 µM, 24.25 µM, 65.97 µM, 175.31 µM, 282.23 µM, 330.29 µM and 369.54 µM, respectively (Li et al., 2021a; Zhou et al., 2021). Sun et al., (2020) found that cyclocarioside Z5 and cyclocarioside Z7 significantly increased glucose consumption in 3 T3-L1 adipocytes, which could be active components for the anti-diabetes effect (Sun et al., 2020). The result and literature reports demonstrated that the ligand fishing strategy based on CFP-immobilized α-glucosidase coupled with UPLC-QTOF-MS/MS could successfully screen the potential α-glucosidase inhibitors from C. paliurus leaves.
3.5 Molecular docking
Molecular docking is an effective method to visualize interactions between ligands and receptors, and predict the possible binding sites and steric conformations of ligands (Álvarez-Chimal et al., 2022; Du et al., 2022). To further elucidate binding interactions of screened potential inhibitors and α-glucosidase, molecular docking was performed. As shown in Table 2 and Fig. 7, Fig. 8, Fig. S5, S6, S7 and S8, cyclocarioside A, cypaliuruside K, cypaliuruside J, cyclocarioside C, cyclocarioside I and pterocaryoside A could smoothly enter the active site pocket of α-glucosidase and interacted with major amino acid residues. From the point view of the affinity, cyclocarioside A, cypaliuruside K, cypaliuruside J exhibited significantly lower binding energy (-9.4, −8.9 and −8.8 kcal/mol, respectively) than the positive control quercetin (-8.5 kcal/mol). Cyclocarioside A interacted with four amino acid residues Gly160, Arg176, Ser162, and Pro151 by forming four hydrogen bonds with the distances ranging from 2.3 Å to 3.7 Å, and formed hydrophobic forces with Pro151, Lys148, Pro149, Trp238 and Phe173 (Fig. 7). Cypaliuruside K formed five hydrogen bonds with amino acid residues Thr310, Arg315, Leu313, Ser240 and His 280, and interacted with Val232, Leu313, Lys156, Arg315, Val216, Phe178 and Tyr158 by hydrophobic forces. The H-bond distances ranged from 2.4 Å to 3.7 Å (Fig. 8). Cypaliuruside J interacted with amino acid residues Ser240, Arg315, Asp307, Pro312, Thr310 and Leu313 by forming seven hydrogen bonds with the average distance of 2.6 Å, and formed hydrophobic bindings with Tyr158, Arg315 and Val216, Phe159 and phe178 (Fig. S5). Although cyclocarioside C, cyclocarioside I and pterocaryoside A had slightly high binding energy (-8.2, −8.1 and 8.0 kcal/mol, respectively), which was close to the affinity of acarbose (-8.1 kcal/mol). Cypaliuruside C might inhibit the α-glucosidase activity by forming four hydrogen bonds with amino acid residues Asp363, Asp362, Gly361 and Ser544, and forming hydrophobic bindings with Lys523, Lys524, Leu323 and Phe321. The H-bond distances ranged from 2.0 Å to 3.5 Å (Fig. S6). As shown in Fig. S7 and Fig. S8, cyclocarioside I and pterocaryoside A formed four hydrogen bonds with amino acid residues Asp363, Asp362 and Phe321, and three hydrogen bonds with Ser241 and Asp242, and their average H-bonds distances were 2.15 Å and 2.33 Å, respectively. They interacted with amino acid residues Leu439 and Phe321 (cyclocarioside I), and with Tyr158, Phe314, and Lys156 (pterocaryoside A) by hydrophobic forces, respectively. Meanwhile, pterocaryoside A interacted with Tyr158 by the pi-sigma interaction. Some studies have reported that amino acid residues Arg 315, Pro312, Tyr158, Pro312, Asp242, Lys156, Ser241, Leu 313, His280, Val216, and Phe178 of α-glucosidase played critical roles in in substrate catalysis and binding mechanism (Xie et al., 2021; Shen et al., 2020; Hu et al., 2021). The results suggested that hydrogen bonds, hydrophobic forces, and Van der Waals were key forces in the binding interactions between potential inhibitors and α-glucosidase. Previous studies demonstrated that hydrogen bonds and hydrophobic interactions between ligands and enzymes could be beneficial to enhance the stability of the inhibitor-enzyme complex, and then exerted essential effects on inhibitory activity toward α-glucosidase (Du et al., 2022; Liu et al., 2021b; Xie et al., 2021).
| Compounds | Affinity (kcal/mol) | Active amino acid residues | Hydrogen bond (Å) | Hydrophobic Interaction (Å) | Pi-Sigma |
|---|---|---|---|---|---|
| Cyclocarioside A | −9.4 | Ala418, Ser162, Arg176, Asn414, Ser180, Thr165, Phe166, Pro149, Phe173, Trp164, Lys148, Pro151, Asp144, Gly161, Ile150, Gly160, Glu421, Thr237, Trp238, Ser157 | Gly160 (2.4), Arg176 (2.5), Ser162 (2.3), Pro151 (3.7) | Alkyl: Pro151 (3.7), Lys148 (3.7), Pro149 (4.6); Pi-Alkyl: Trp238 (4.7, 5.4), Phe173 (4.8, 5.3) | – |
| Cypaliuruside K | −8.9 | Ser240, Val232, Leu313, Asp233, Pro312, Thr310, Asp307, Phe314, Ser311, Phe178, His280, Val216, Arg442, Glu277, Asp352, Gln279, Phe303, Glue411, Tyr158, Lys156, Ser157, Asp242, Arg315 | Thr310 (2.6), Arg 315 (2.4), Leu313 (3.7), Ser 240 (2.8), His 280 (3.0) | Alkyl: Val232 (5.1), Leu 313 (4.5), Lys156 (4.1, 5.4), Arg 315 (3.8, 5.3); Pi-Alkyl: Val216 (4.8, 5.3), Phe178 (4.7, 5.4), Tyr158 (3.5, 4.2, 4.6, 5.2) | – |
| Cypaliuruside J | −8.8 | Leu313, Phe314, Arg315, Asp307, Ser311, Pro312, Thr310, Phe159, His280, Asp352, Phe178, Val216, Gln279, Arg442, Glu277, Phe303, Glu411, Tyr158, Lys156, Ser157, Asp242, Ser240 | Ser240 (2.5), Arg315 (2.2), Asp307 (2.6), Thr310 (2.6), Pro312(2.0,3.1), Leu313 (3.5) | Alkyl: Tyr158 (3.4, 4.0, 4.2, 5.1), Arg315 (3.7, 5.2), Val216 (4.8); Pi-Alkyl: Phe159 (4.9), Phe178 (4.4, 5.4) | – |
| Cyclocarioside C | −8.2 | Lys523, Lys524, Leu323, Glu322, Phe543, Phe321, Arg359, Gly361, Asp362, Ser364, Asp363, Leu439, Lys400, Thr358, Trp581, Ser545, Asp546, Ser544 | Asp363 (2.1), Asp362 (2.0), Gly361 (2.8), Ser544 (3.5) | Alkyl: Lys523 (4.2, 4.3, 5.1), Lys524 (4.4), Leu323 (4.6); Pi-Alkyl: Phe321(5.0) | – |
| Cyclocarioside I | −8.1 | Lys400, Thr358, Asp363, Ile357, Asp362, Ser364, Gly361, Phe360, Arg359, Trp581, Leu323, Ser544, Lys524, Lys523, Phe543, Phe321, Glu322, Pro320, Leu439 | Asp363 (2.0, 2.4), Asp362 (2.0), Phe321 (2.2) |
Alkyl: Leu439(4.4, 5.2) Pi-Alkyl: Phe321(4.5, 5.1) |
– |
| Pterocaryoside A | −8.0 | Asp242, Ser241, Arg315, Thr245, Phe303, Gln279, Asp307, Leu246, His280, Pro312, Leu313, Ser240, Lys156, Phe314, Tyr158, Leu177 | Ser241 (2.1, 2.7), Asp242 (2.2) |
Alkyl: Lys156 (4.7) Pi-Alkyl: Tyr158 (5.1), Phe314 (4.8) |
Tyr158 (3.5) |
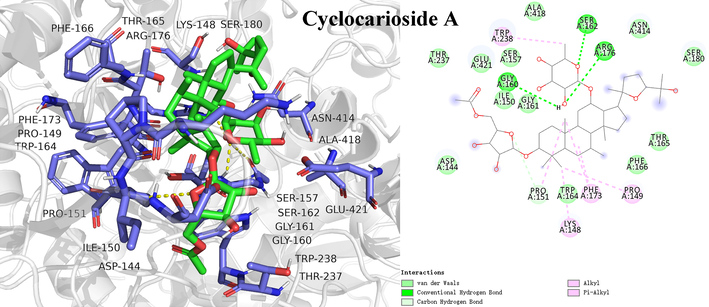
- Molecular docking study of potential inhibitor cyclocarioside A interacting with amino acid residues in the active site of α-glucosidase.
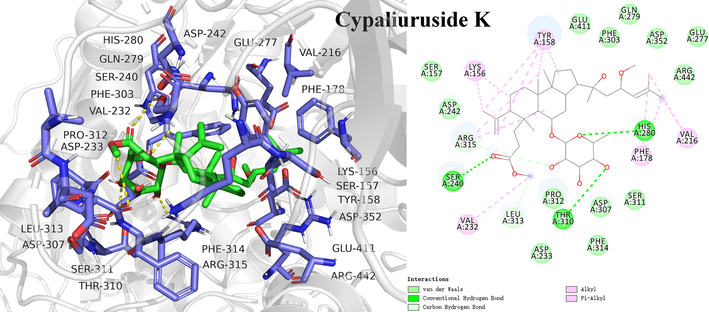
- Molecular docking study of the potential inhibitor cypaliuruside K interacting with amino acid residues in the active site of α-glucosidase.
4 Conclusions
In this study, a target enzyme-oriented fishing tool of CFP-immobilized α-glucosidase combined with UPLC-QTOF-MS/MS was developed to fish out and identify potential α-glucosidase inhibitors from C. paliurus leaves. A total of 36 potential α-glucosidase inhibitors were successfully screened, and further identified by UPLC-QTOF-MS/MS. Moreover, inhibitory activities of screened active compounds against α-glucosidase were evaluated by enzyme inhibitory assay in vitro. Molecular docking further illustrated inhibitory mechanisms between potential inhibitors and α-glucosidase. Docking results showed that cyclocarioside A, cypaliuruside K, cypaliuruside J, cyclocarioside C, cyclocarioside I and pterocaryoside A could embed into the active pocket of the model, and mainly interacted with critical amino acid residues by forming hydrogen bonds, hydrophobic forces, and Van der Waals. Results demonstrated that the target enzyme-oriented ligand fishing method is effective and reliable to capture potential α-glucosidase inhibitors from complex mixtures.
Acknowledgments
This work was supported by the Special Foundation for National Science and Technology Basic Resources of China [2018FY100701]; the Key Program of Natural Science Foundation of Gansu Province, China [20JR10RA584]; the 2021 project of State Drug Administration-Key Laboratory of Quality Control of Chinese Medicinal Materials and Decoction Pieces, China [2021GSMPA-KL12]; and the Education Department of Gansu Province: Excellent Graduate Student “Innovation Star” Project [2021CXZX-019].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Development of diarylpentadienone analogues as alpha-glucosidase inhibitor: synthesis, in vitro biological and in vivo toxicity evaluations, and molecular docking analysis. Bioorg. Chem.. 2020;104:104277
- [Google Scholar]
- Influence of the particle size on the antibacterial activity of green synthesized zinc oxide nanoparticles using Dysphania ambrosioides extract, supported by molecular docking analysis. Arab. J. Chem.. 2022;15(6):103804
- [Google Scholar]
- Phenolic profiles and screening of potential α-glucosidase inhibitors from Polygonum aviculare L. leaves using ultra-filtration combined with HPLC-ESIqTOF-MS/MS and molecular docking analysis. Ind. Crop. Prod.. 2020;154:112673
- [Google Scholar]
- Dual-target screening of bioactive components from traditional Chinese medicines by hollow fiber-based ligand fishing combined with liquid chromatography-mass spectrometry. J. Pharmaceut. Biomed.. 2017;143:269-276.
- [Google Scholar]
- Qingqianliusus A-N, 3,4-seco-dammarane triterpenoids from the leaves of Cyclocarya paliurus and their biological activities. Arab. J. Chem.. 2023;16(1):104441
- [Google Scholar]
- New triterpenoid saponins from the leaves of Cyclocarya paliurus. Chinese Chem. Lett.. 2015;26:585-589.
- [Google Scholar]
- C-methyl flavonoid from the leaves of Cleistocalyx conspersipunctatus: α-glucosidase inhibitory, molecular docking simulation and biosynthetic pathway. Arab. J. Chem.. 2022;15(4):103687
- [Google Scholar]
- Triterpenoids from Cyclocarya paliurus that enhance glucose uptake in 3T3-L1 adipocytes. Molecules. 2019;24:187.
- [Google Scholar]
- Essential moieties of myricetins, quercetins and catechins for binding and inhibitory activity against α-Glucosidase. Bioorg. Chem.. 2021;115:105235
- [Google Scholar]
- α-Glucosidase inhibitors screening from Cyclocarya paliurus based on spectrum-effect relationship and UPLC-MS/MS. Biomed. Chromatogr.. 2022;36:5313.
- [Google Scholar]
- Recent advances in screening active components from natural products based on bioaffinity techniques. Acta Pharm. Sin. B. 2020;10(10):1800-1813.
- [Google Scholar]
- New ent-kaurane diterpenes from the roasted arabica coffee beans and molecular docking to α-glucosidase. Food Chem.. 2021;345:128823
- [Google Scholar]
- Computational designing of low energy gap small molecule acceptors for organic solar cells. J. Mex. Chem. Soc.. 2017;61(4):309-316.
- [Google Scholar]
- Two new triterpenoid glycosides from Cyclocarya paliurus. J. Asian Nat. Prod. Res.. 2006;8(1–2):93-98.
- [Google Scholar]
- Novel highly sweet secodammarane glycosides from pterocarya paliurus. J. Agr. Food Chem.. 1995;43(10):2602-2607.
- [Google Scholar]
- Hypoglycemic action of Cyclocarya paliurus (Batal.) Iljinskaja in normal and diabetic mice. Biosci. Biotechnol. Biochem.. 2003;67(4):877-880.
- [Google Scholar]
- New triterpenoids from the leaves of Cyclocarya paliurus. Planta Med.. 2012;78:290-296.
- [Google Scholar]
- α-Glucosidase inhibitory and anti-inflammatory activities of dammarane triterpenoids from the leaves of Cyclocarya paliurus. Bioorg. Chem.. 2021;111:104847
- [Google Scholar]
- Dopamine-polyethyleneimine co-deposition cellulose filter paper for α-Glucosidase immobilization and enzyme inhibitor screening. J. Chromatogr. B. 2021;1167:122582
- [Google Scholar]
- Screening and identification of α-glucosidase inhibitors from Cyclocarya paliurus leaves by ultrafiltration coupled with liquid chromatography-mass spectrometry and molecular docking. J. Chromatogr. A. 2022;1675:463160
- [Google Scholar]
- Inhibitory mechanism of sinensetin on α-glucosidase and non-enzymatic glycation: insights from spectroscopy and molecular docking analyses. Int. J. Biol. Macromol.. 2021;166:259-267.
- [Google Scholar]
- α-Glucosidase immobilization on chitosan-modified cellulose filter paper: preparation, property and application. Int. J. Biol. Macromol.. 2019;122:298-305.
- [Google Scholar]
- 3,4-seco-Dammarane Triterpenoid Saponins with anti-inflammatory activity isolated from the leaves of Cyclocarya paliurus. J. Agr. Food Chem.. 2020;68:2041-2053.
- [Google Scholar]
- A new functionality study of vanillin as the inhibitor for α-glucosidase and its inhibition kinetic mechanism. Food Chem.. 2021;353(7):129448
- [Google Scholar]
- Theoretical investigation for the designing of novel antioxidants. Can. J. Chem.. 2013;91:126-130.
- [Google Scholar]
- Quantum chemical designing of novel organic non-linear optical compounds. Bull. Korean Chem. Soc.. 2014;35:1391.
- [Google Scholar]
- Recent progress in porphyrin-based materials for organic solar cells. J. Mater. Chem. A. 2018;6:16769.
- [Google Scholar]
- Identification of α-glucosidase inhibitors from cyclocarya paliurus tea leaves using UF-UPLC-QTOF-MSMS and molecular docking. Food Funct.. 2019;10(4):1893-1902.
- [Google Scholar]
- Qin, J.J., 2018. Study on chemical constituents of Periploca chrysantha, Cynanchum bungei, Cyclocarya paliurus, and Celastrus monospermus and evaluation of their biological activities. PhD thesis, Shanghai Institute of Materia Medica, University of Chinese Academy of Sciences, Shanghai, 77-118
- Construction of Fe4O3@α-glucosidase magnetic nanoparticles for ligand fishing of α-glucosidase inhibitors from a natural tonic Epimedii Folium. Int. J. Biol. Macromol.. 2020;165:1361-1372.
- [Google Scholar]
- Studies on the sweet principles from the leaves of Cyclocarya paliurus (Batal.) Iljinsk. Acta Pharm. Sin. B. 1995;30:757-761.
- [Google Scholar]
- Hypoglycemic triterpenoid glycosides from cyclocarya paliurus (sweet tea tree) Bioorg. Chem.. 2020;95:103493
- [Google Scholar]
- α-glucosidase immobilization on magnetic core-shell metal-organic frameworks for inhibitor screening from traditional Chinese medicines. Colloid Surf. B. 2021;205:111847
- [Google Scholar]
- New dammarane triterpenoid saponins from the leaves of Cyclocarya paliurus. J. Asian Nat. Prod. Res.. 2018;20(11):1019-1027.
- [Google Scholar]
- A strategy for screening of α-glucosidase inhibitors from Morus alba root bark based on the ligand fishing combined with high-performance liquid chromatography mass spectrometer and molecular docking. Talanta. 2018;180:337-345.
- [Google Scholar]
- Fatigue-alleviating effect of polysaccharides from Cyclocarya paliurus (Batal) Iljinskaja in mice. Afr. J. Microbiol. Res.. 2012;6(24):5243-5248.
- [Google Scholar]
- Inhibition mechanism of α-glucosidase inhibitors screened from Artemisia selengensis Turcz root. Ind. Crop. Prod.. 2020;143:111941
- [Google Scholar]
- Chemical constituents from Cyclocarya paliurus (Batal.) Iljinsk. Biochem. Syst, Ecol.. 2014;57:216-220.
- [Google Scholar]
- Triterpenoids from Cyclocarya paliurus and their inhibitory effect on the secretion of apoliprotein B48 in Caco-2 cells. Phytochemistry. 2017;142:76-84.
- [Google Scholar]
- Combined magnetic ligand fishing and high-resolution inhibition profiling for identification of α-glucosidase inhibitory ligands: a new screening approach based on complementary inhibition and affinity profiles. Talanta. 2019;200:279-287.
- [Google Scholar]
- Screening α-glucosidase inhibitors from four edible brown seaweed extracts by ultra-filtration and molecular docking. LWT - Food Sci. Technol.. 2021;138:110654
- [Google Scholar]
- Sulfated modification, characterization and antioxidant activities of polysaccharide from Cyclocarya paliurus. Food Hydrocolloid. 2016;53:7-15.
- [Google Scholar]
- Phytochemicals from the Leaves of Cyclocarya paliurus and their 11β-HSD1 enzyme inhibitory effects. Chem. Biodivers.. 2021;18:e2000772.
- [Google Scholar]
- Cyclocarya paliurus polysaccharides alleviate type 2 diabetic symptoms by modulating gut microbiota and short-chain fatty acids. Phytomedicine. 2020;77:153268
- [Google Scholar]
- Phenolic compounds from the leaves of Cyclocarya paliurus (Batal.) Ijinskaja and their inhibitory activity against PTP1B. Food Chem.. 2010;119:1491-1496.
- [Google Scholar]
- Screening acetylcholinesterase inhibitors from traditional Chinese medicines by paper-immobilized enzyme combined with capillary electrophoresis analysis. J. Pharmaceut. Biomed.. 2020;190:113547
- [Google Scholar]
- Effect of Cyclocarya Paliurus on hypoglycemic effect in type 2 diabetic mice. Med. Sci. Monitor. 2019;25:2976-2983.
- [Google Scholar]
- Bioactive dammarane triterpenoid saponins from the leaves of cyclocarya paliurus. Phytochemistry. 2021;183:112618
- [Google Scholar]
- Four new dammarane triterpenoid glycosides from the leaves of Cyclocarya paliurus and their SIRT1 activation activities. Fitoterapia. 2021;154:105003
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.104802.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







