Translate this page into:
Design, synthesis, and biological evaluation of (E)-N′-substitute-4-((4-pyridylpyrimidin-2-yl)amino)benzohydrazide derivatives as novel potential CDK9 inhibitors
⁎Corresponding author at: Meijuan Fang, School of Pharmaceutical Sciences, Xiamen University, South Xiang-An Road, Xiamen 361102, China. fangmj@xmu.edu.cn (Meijuan Fang)
⁎⁎Corresponding author at: Xingzhi College, Zhejiang Normal University, Lanxi 321004, China. huhongyu22@126.com (Hongyu Hu),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
CDK9 is a promising drug target for treating many diseases such as cancer and HIV. In our previous studies, two series of 5-(substituted amino)-1H-indole-2-carbohydrazide derivatives were discovered as CDK9 inhibitors exhibiting potent HIV-1 transcription inhibition and anticancer activities. In a continuing effort to develop new CDK9 inhibitors endowed with good anticancer activity, we designed and synthesized a series of new benzene carbohydrazide derivatives bearing a (pyridyl pyrimidin-2-yl)amino moiety in position-4 of the phenyl ring. This work reports the preparation of benzene carbohydrazide derivatives, their inhibition effect on HIV-1 transcription, and the preliminary structure–activity relationships. Compound 9h was found to be the most potent CDK9 inhibitor and exhibited excellent anti-proliferative activities against cancer cells (A375, A549, HepG2, and MCF-7) but was less toxic to normal cells (MCF-10A and HaCaT). Further bioassays indicated that compound 9h could induce the apoptosis of cancer cells, which contributes to its antitumor effects. Finally, we performed molecular docking studies to predict the binding mode of 9h at the ATP binding site of CDK9 and identify the essential amino acids responsible for the interactions.
Keywords
Benzene carbohydrazide derivatives
CDK9
HIV-1 transcription inhibition activity
Anticancer
Structure-activity relationship (SAR)
Apoptosis
1 Introduction
Cyclin-dependent kinases (CDKs) are a family of serine/threonine kinases that control cell cycle and cell transcription (Doree and Galas 1994, Malumbres 2014). As a serine kinase, CDK9 and its partner Cyclin T comprise positive transcription elongation factor b (P-TEFb), which plays a crucial role in transcriptional regulation (Peng et al., 1998). P-TEFb phosphorylates Polymerase II (Pol II) C-terminal domain (CTD) Serine-2, which subsequently recruits other transcription factors or cofactors for productive transcription. Moreover, P-TEFb also phosphorylates the subunit of DRB Sensitivity Inducing Factor (DSIF) and negative elongation factor (NELF), which inhibits the negative effect of NELF and converts DSIF into a positive elongation factor (Zhou et al., 2012). Pol II pause release is a rate-limiting step for transcription elongation, especially for HIV and cancers (Rahl et al., 2010, Lu et al., 2013, Huang et al., 2014). Therefore, CDK9 is a valuable target for antiviral and antitumor drug development. Previous studies have shown that CDK9 inhibitors potently induced cancer cell apoptosis by inhibiting the activity of Pol II and reducing intracellular mRNA levels of various anti-apoptotic proteins (Rahaman et al., 2019).
At present, some small molecules targeting CDK9 have been reported, including selective and pan-CDK inhibitors. As shown in Fig. 1, Flavopiridol (FLP), one of the earliest nonselective CDKs inhibitors, was the first CDK9 inhibitor to enter clinical trials (Senderowicz 1999) for the treatment of chronic lymphocytic leukemia (CLL) (Kouroukis et al., 2003). However, the poor selectivity and side effects limited its further development. After that, several targeting CDK9 inhibitors, including LS-007 (Walsby et al., 2014, Xie et al., 2016), Atuveciclib (Lücking et al., 2017), ly2857785 (Yin et al., 2014), and NVP-2 (Olson et al., 2018) were subsequently discovered. Some of them have entered into clinical trials. For instance, LS-007 exhibited potent inhibition over CDK1, CDK2, CDK4, and CDK9 with IC50 in the range of 3–8 nM, which can treat acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML). ly2857785, a potent CDK9 inhibitor, inhibited the growth of a wide range of cancer cell lines, especially in hematologic tumor cells. Atuveciclib and NVP-2 also showed high potency against CDK9 to treat multi-malignancies. Albeit numerous prospective targeting CDK9 molecules have been developed and validated, there were no reported inhibitors that solve the problems of pan-CDK inhibitors with low selectivity and high toxicity properties (Wu et al., 2020). Therefore, it is necessary to continually discover new targeting CDK9 entities with excellent biological activities and physicochemical properties.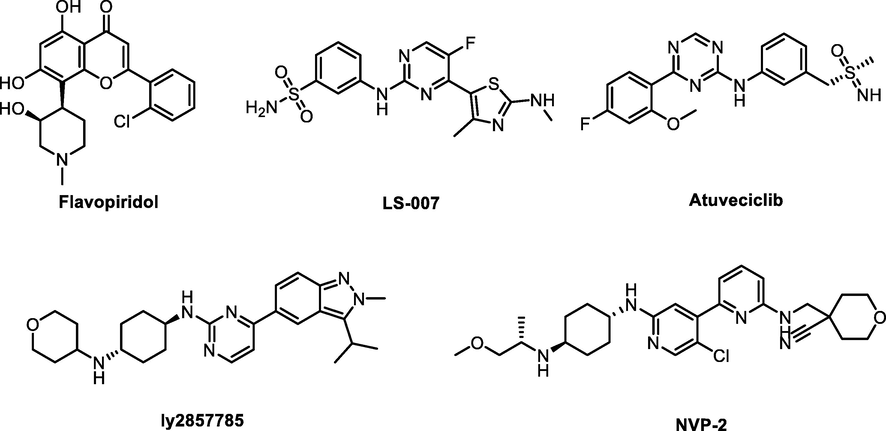
Structures of representative CDK9 inhibitors.
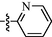
Our previous research found that 2,5-disubstituted indole derivative 3b could act as a CDK9 inhibitor to impede the phosphorylation at Ser2 of RNAPII CTD and induce cancer cells apoptosis (Hu et al., 2016). Subsequently, compound 12i derived from 3b exerted significant inhibition of CDK9 activity and excellent anticancer activity but low toxic activity on normal cells (Hu et al., 2020). Generally, the pyrimidin-2-amine-4-yl moiety contained in many kinase inhibitors helps suppress kinase activity. It can provide hydrogen bond donors and hydrogen bond acceptors to form hydrogen bond interactions with key residues in the hinge region of the kinases. This study wanted to further optimize 12i using scaffold hopping and substitution decorating strategies (Fig. 2). For lead compound 12i, we retained 2-(λ2azanyl)-4-(pyridin-3-yl)pyrimidine moiety and carbohydrazide group, changed the indole scaffold to a phenyl ring, and introduced different substitutions at the position of the methylene group (R1), building the structure of the target compounds 9a-9n (Fig. 2). To evaluate the effect of the pyridine link position on the inhibitory activity, the m-pyridine on the amino was then replaced by o- and p-pyridine to get compounds 10a-10n and 11a-11n, respectively.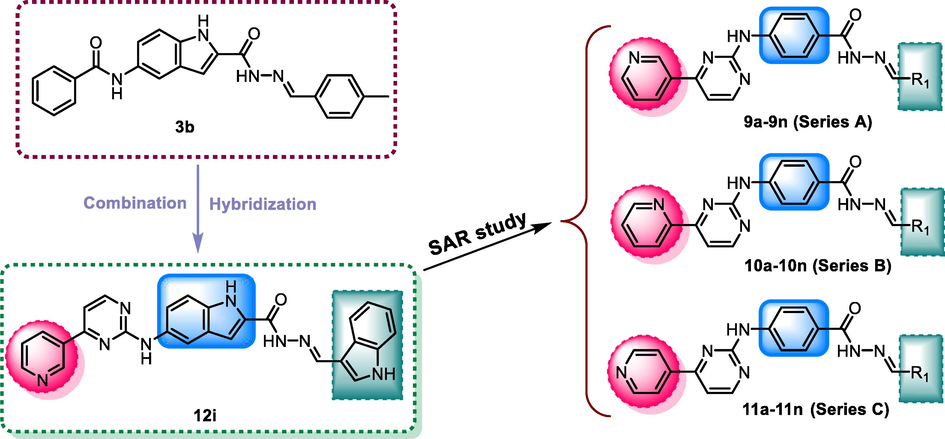
The design strategy of compounds 9a-9n, 10a-10n, and 11a-11n.
In this study, we synthesized three series target compounds and evaluated their CDK9 inhibitory by using a luciferase transcription assay for HIV-1 transcription efficiency. From the preliminary studies of the CDK9 inhibitory, active compounds were further taken up for screening anticancer activity against a panel of four cancer cell lines and the toxic effect on two normal cell lines. Subsequently, the most active compound, 9h, was evaluated to study its effect on inhibition of phosphorylation of the RNAPII CTD Ser2, the expression levels of cleaved PARP (apoptosis-related protein), and induction of cell apoptosis. Finally, we performed molecular docking studies to predict the possible binding model of 9h and CDK9.
2 Results and discussion
2.1 Chemistry
The synthetic route of the target compounds is outlined in Scheme 1. The refluxing of ethyl 4-aminobenzoate (1) and cyanamide in the presence of concentrated HCl yielded ethyl 4-guanidinobenzoate (2). Compound 2 refluxed with 3-dimethylamino-1-(3-pyridyl)-2-propen-1-one, 3-dimethylamino-1-(2-pyridyl)-2-propen-1-one, and 3-dimethylamino-1-(4-pyridyl)-2-propen-1-one afforded 3, 4, and 5 respectively. Then, compounds 3–5 reacted with hydrazine hydrate to form hydrazine products 6–8, respectively. The further condensation reaction of hydrazine products 6–8 with the appropriate aldehydes gave the corresponding hydrazone products 9a-9n, 10a-10n, and 11a-11n, respectively. The structures of the target compounds were characterized by 1H NMR, 13C NMR, and HRMS.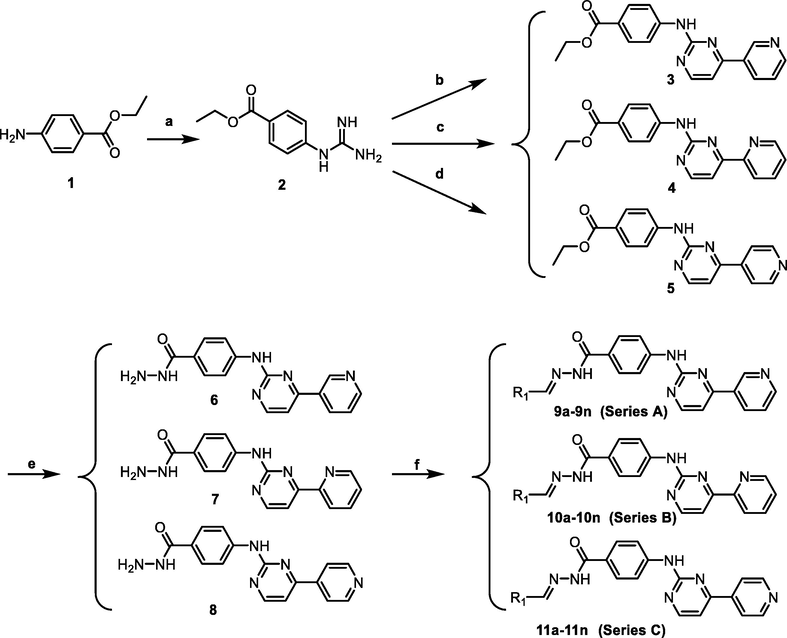
The synthetic route of target compounds. Reagents and conditions: (a) H2NCN, concd HCl, EtOH, reflux, 24 h, NH4NO3 (aq); (b) 3-(dimethylamino)-1-(3-pyridinyl)prop-2-en-1-one, NaOH, EtOH, reflux, 48 h; (c) (E)-3-(dimethylamino)-1-(pyridin-2-yl)prop-2-en-1-one, NaOH, EtOH, reflux, 48 h; (d) (E)-3-(dimethylamino)-1-(pyridin-4-yl)prop-2-en-1-one, NaOH, EtOH, reflux, 48 h; (e) NH2NH2·H2O, 95% EtOH, 80 °C, 8 h; (f) RCHO, EtOH, 80 °C, 2 h.
2.2 Inhibition of CDK9 activity and SAR analysis
CDK9 is a core kinase of the P-TEFb complex and regulates transcription elongation, especially for HIV-1, oncogenes, and cancer suppressors. NH2 cells obtain an integrated HIV-LTR-luciferase reporter gene and Tat-Flag, which simulates the CDK9-dependent HIV-1 transcription (Li et al., 2013). As CDK9 inhibitors could effectively inhibit HIV-1 transcription, we used the HIV-1 transcription cell model NH2 for screening to intuitively and rapidly detect the inhibitory of target compounds (Hu et al., 2016; Hu et al., 2020). By comparing the luciferase expression in compound-treated NH2 cells with that in DMSO-treated NH2 cells, we calculated the HIV-1 transcription activity of NH2 cells treated with each compound at three different concentrations (Tables 1–3). The low rate of HIV-1 transcription activity means a high potential inhibition of CDK9. Among these three series of target compounds, we found that nine compounds at 20 µM exhibited a good inhibition of CDK9 activity with the HIV-1 transcription rate of less than 20%. Compound 9h was identified as the most potential CDK9 inhibitor because it inhibited 96% HIV-1 transcription in NH2 cells. These results showed the relationships between the inhibitory activity and structure of target compounds, and SARs analyses were discussed as follows. The values are the mean ± SD of at least three independent experiments. NT: Not Tested. The values are the mean ± SD of at least three independent experiments. NT: Not Tested. The values are the mean ± SD of at least three independent experiments. NT: Not Tested.
Compound
Structure
HIV-1 transcription activity
R1
0.2 (μM)
2.0 (μM)
20 (μM)
9a

0.82 ± 0. 10
0.77 ± 0.02
0.41 ± 0.05
9b

0.80 ± 0.01
0.73 ± 0.10
0.51 ± 0.02
9c
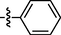
0.77 ± 0.02
0.50 ± 0.04
0.21 ± 0.01
9d
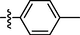
0.47 ± 0.05
0.38 ± 0.08
0.32 ± 0.05
9e
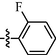
1.00 ± 0.10
0.44 ± 0.04
0.30 ± 0.01
9f
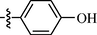
0.77 ± 0.10
0.55 ± 0.12
0.15 ± 0.04
9g
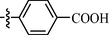
0.85 ± 0.20
0.77 ± 0.06
0.67 ± 0.10
9h
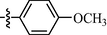
0.70±0.10
0.26±0.03
0.04±0.01
9i
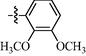
0.93 ± 0.08
0.59 ± 0.06
0.27 ± 0.02
9j
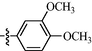
0.81 ± 0.05
0.39 ± 0.01
0.17 ± 0.02
9k
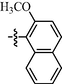
0.87 ± 0.02
0.57 ± 0.01
0.45 ± 0.02
9l
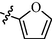
0.79 ± 0.09
0.54 ± 0.03
0.13 ± 0.01
9m
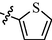
0.78 ± 0.02
0.56 ± 0.03
0.28 ± 0.01
9n
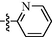
0.87 ± 0.02
0.61 ± 0.03
0.16 ± 0.05
FLP
0.15 ± 0.05
NT
NT
3b
0.76 ± 0.06
0.64 ± 0.05
0.26 ± 0.03
12i
0.2 ± 0.10
0.09 ± 0.08
0.07 ± 0.03
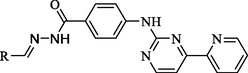
Compound
Structure
HIV-1 transcription activity
R1
0.2 (μM)
2.0 (μM)
20 (μM)
10a

0.83 ± 0.15
0.69 ± 0.10
0.36 ± 0.01
10b

0.75 ± 0.10
0.63 ± 0.05
0.32 ± 0.07
10c
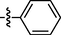
1.00 ± 0.18
0.34 ± 0.05
0.22 ± 0.02
10d
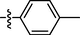
0.85 ± 0.10
0.57 ± 0.10
0.43 ± 0.05
10e
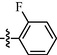
0.81 ± 0.06
0.46 ± 0.05
0.24 ± 0.02
10f
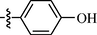
0.82 ± 0.09
0.73 ± 0.15
0.22 ± 0.02
10g
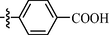
0.85 ± 0.09
0.84 ± 0.06
0.78 ± 0.04
10h
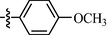
0.87 ± 0.05
0.46 ± 0.05
0.14 ± 0.02
10i
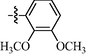
1.00 ± 0.15
0.60 ± 0.14
0.27 ± 0.06
10j
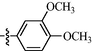
0.74 ± 0.08
0.39 ± 0.05
0.18 ± 0.10
10k
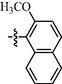
1.00 ± 0.20
0.92 ± 0.10
0.78 ± 0.12
10l
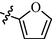
0.89 ± 0.12
0.50 ± 0.06
0.07 ± 0.01
10m
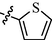
1.00 ± 0.25
0.50 ± 0.14
0.27 ± 0.07
10n
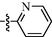
0.87 ± 0.09
0.61 ± 0.05
0.33 ± 0.01
FLP
Positive compounds
0.15 ± 0.05
NT
NT
3b
0.76 ± 0.06
0.64 ± 0.05
0.26 ± 0.03
12i
0.2 ± 0.10
0.09 ± 0.08
0.07 ± 0.03
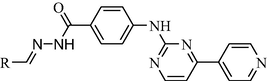
Compound
Structure
HIV-1 transcription activity
R1
0.2 (μM)
2.0 (μM)
20 (μM)
11a

0.92 ± 0.12
0.87 ± 0.08
0.87 ± 0.20
11b

0.97 ± 0.22
0.73 ± 0.12
0.71 ± 0.09
11c
0.85 ± 0.11
0.56 ± 0.08
0.45 ± 0.08
11d
0.91 ± 0.12
0.69 ± 0.09
0.30 ± 0.02
11e
0.93 ± 0.12
0.64 ± 0.08
0.11 ± 0.02
11f
0.97 ± 0.22
0.75 ± 0.12
0.45 ± 0.10
11g
0.90 ± 0.12
0.89 ± 0.05
0.72 ± 0.12
11h
0.76 ± 0.10
0.36 ± 0.08
0.24 ± 0.04
11i

0.86 ± 0.19
0.67 ± 0.05
0.36 ± 0.02
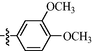
11j
0.96 ± 0.15
0.67 ± 0.10
0.43 ± 0.08
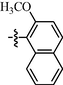
11k
0.98 ± 0.10
0.87 ± 0.19
0.24 ± 0.02
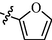
11l
0.71 ± 0.10
0.66 ± 0.12
0.33 ± 0.05
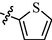
11m
0.77 ± 0.07
0.73 ± 0.10
0.28 ± 0.06
11n
1.00 ± 0.18
0.76 ± 0.23
0.47 ± 0.08
FLP
Positive compounds
0.15 ± 0.05
NT
NT
3b
0.76 ± 0.06
0.64 ± 0.05
0.26 ± 0.03
12i
0.2 ± 0.10
0.09 ± 0.08
0.07 ± 0.03
2.2.1 N′-methylene-4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzohydrazide Derivatives (9a-9n)
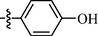
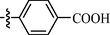
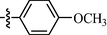
As shown in Table 1, all of the N′-methylene-4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzohydrazide derivatives (9a-9n, Series A) possessed a different degree of inhibitory ability of the HIV-1 transcription. When R1 was an aliphatic alkyl group, the inhibition of the HIV-1 transcription effect was weak. We found that both 9a (R1 = butyl group) and 9b (R1 = pentyl group) showed the inhibitory activity with the HIV-1 transcription inhibition rates of 59% and 49% at 20 µM, respectively. When R1 was an aromatic group, almost all compounds (9c-9n, except 9g) exhibited >50% HIV-1 transcriptional inhibitory activity. Compound 9g bearing a p-carboxylphenyl group at the position of the methylene group displayed low activity to inhibit HIV-1 transcription with inhibition rates of 23% at 2.0 µM and 33% at 20 µM, respectively, implying that an acid electron-withdrawing substituent on the aromatic ring greatly weakened the inhibitory activity. Compound 9k with a naphthalene moiety at the position of the methylene group also had lower inhibitory activity (HIV-1 transcriptional inhibition rates of 43% at 2.0 µM and 55% at 20 µM). Compounds 9l-9n with an aromatic heterocyclic substitution had similar inhibitory activities with compounds with unsubstituted (9c) or substituted (9d-9f and 9h-9j) phenyl ring. They had the inhibitory effects at 20 µM in the order 2-furyl (9l) > 2-pyridyl (9n) > 2-thienyl (9m). Among compounds 9c-9j with R1 of a mono- or di-substituted benzene ring, compound 9h (R1 = p-methoxyphenyl group) displayed the most potent HIV-1 transcription inhibitory activity with inhibition rates of 74% and 96% at 2.0 µM and 20 µM, respectively.
2.2.1.1 N′-methylene-4-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzohydrazide Derivatives (10a-10n, series B)
Among target compounds with 2-pyridyl on the 4-position of pyrimidine (Table 2), we found that 10a (64% at 20 µM) and 10b (68% at 20 µM) with aliphatic alkyl groups had a higher inhibitory potency on HIV-1 transcription than corresponding 9a (59% at 20 µM) and 9b (49% at 20 µM) but still less active than most of the compounds with an aromatic group (10c-10n). Besides, like 9g and 9k, compounds with a p-carboxylphenyl group (10g: 22% at 20 µM) and a naphthalene moiety (10k: 22% at 20 µM) displayed similar less inhibitory activity than other N′-methylene-4-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzohydrazide derivatives. In comparison, compounds (10l-10n) with an aromatic heterocyclic group possessed similar or better inhibitory activities than compounds (10c-10j) with unsubstituted or substituted phenyl groups. For instance, 10l (R1 = 2-furyl, 93% at 20 µM) showed the best HIV-1 transcription inhibition among compounds 10c-10n. In contrast, compounds (10c-10j) with an unsubstituted or substituted phenyl ring had inhibitory activity rates from 57% to 86% at 20 µM. In particular, compound 10h with R1 of the p-methoxyphenyl group also exhibited the most potent inhibitory effect (86% at 20 µM) in these phenyl substituted derivatives (10c-10j). The results suggested that the SAR of N′-methylene-4-((4-(pyridin-2-yl)pyrimidin-2-yl)amino)benzohydrazide derivatives was consistent with that of the N′-methylene-4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzohydrazide derivatives.
2.2.1.2 N′-methylene-4-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)benzohydrazide Derivatives (11a-11n, series C)
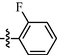
For compounds with 3-pyridyl on the 4-position of pyrimidine (Table 3), we found compound 11e (89% at 20 µM) with an o-fluoro phenyl group had the best activity. In comparison, compound 11h (76% at 20 µM) bearing a p-methoxyphenyl group only had moderate inhibitory activity. Among series C of target compounds, compounds 11a (13% at 20 µM) and 11b (29% at 20 µM) with aliphatic groups displayed lower inhibitory potency on HIV-1 transcription than those with R1 of aromatic moieties (53–89% at 20 µM, except 11g). Among compounds with aromatic groups (11c-11n), 11g with a p-carboxylphenyl group exhibited the lowest inhibitory activity. These results are consistent with those of the above two series of target compounds. Compared with the corresponding compounds of series A (9c-9f, 9h-9j, and 9l-9n) and series B (10c-10f, 10h-10j, and 10l-10n), most of the target compounds of the series C (11c-11f, 11h-11j, and 11l-11n) displayed lower inhibitory activities. However, when R1 was of a 2-methoxynaphthalen-1-yl group, 11k (76% at 20 µM) showed a better inhibition than 9k (55% at 20 µM) and 10k (22% at 20 µM). Compound 11e (89% at 20 µM) with R1 of an o-fluoro phenyl group had higher activity than the corresponding 9e (70% at 20 µM) and 10e (76% at 20 µM).
Overall, we could conclude that both the substituent (R1) on the carbon of the methylene group and the pyridyl group on the C-4 position of the pyrimidine ring could significantly affect the inhibitory activity of HIV-1 transcription. For substituent R1, aromatic-substituted derivatives generally have better inhibitory activity than aliphatic hydrocarbon-substituted compounds. Introducing an acid electron-withdrawing group (—COOH) to the phenyl ring was unfavorable to the inhibitory activity of HIV-1 transcription. Apart from that, the effect of the N-position of the pyridyl ring on the inhibitory activity of HIV-1 transcription was 3-pyridyl > 2-pyridyl > 4-pyridyl. The SARs of synthesized compounds are summarized in Fig. 3. Additionally, the active compound 9h displayed better CDK9 inhibition than our previously reported compound 3b; however, compared with our another reported compound 12i, 9h exhibited similar activity at 20 μM but less inhibitory activity at low concentrations (0.2 μM and 2.0 μM). Both 12i and 9h bear the 4-(pyridin-3-yl)pyrimidin-2-yl substitution at their parent nucleus (C5-position of indole ring and C4-position of benzene ring, repectively) and the 4-(pyridin-3-yl)pyrimidin-2-yl moiety might be a good pharmacophore which can help to form the optimal interactions of a small molecule with CDK9 biologic target and to block its biologic response (HIV-1 transcription).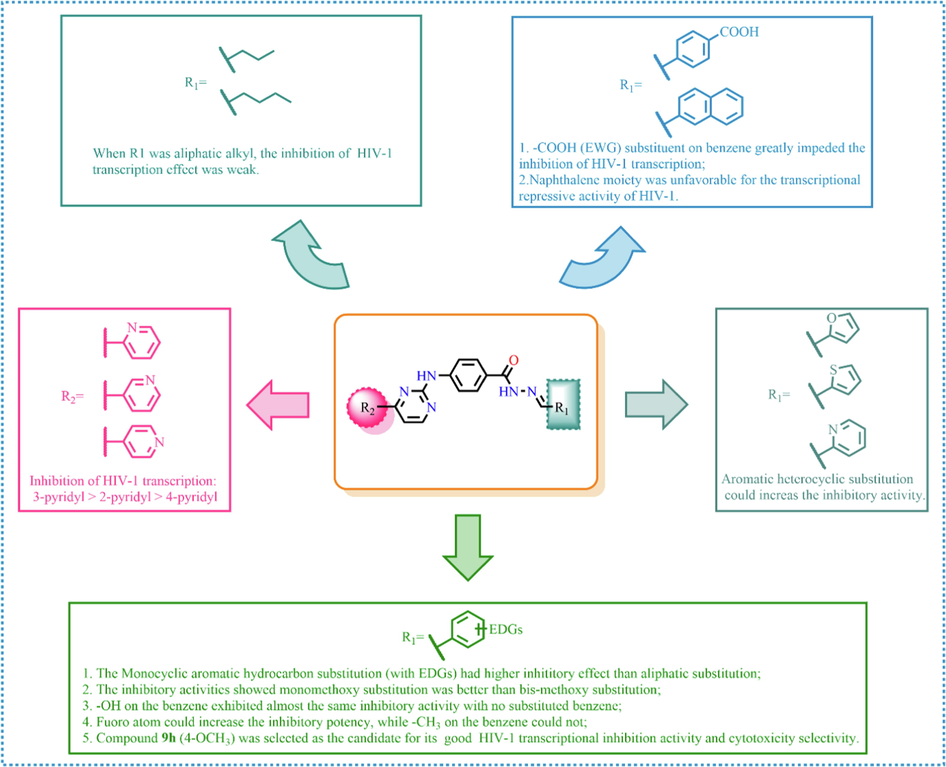
Summary of the structure–activity relationship.
2.3 Cell proliferation evaluation of selected compounds against cancer and normal cells
Based on the results of the SAR analysis, the most active compounds at 20 µM, including 9f, 9h, 9j, 9l, 10h, 10j and 10l, were selected as seed compounds for further evaluation of the anti-proliferative activities against four cancer cell lines (A375, A549, HepG2, and MCF-7) and two normal cell lines (Hacat and MCF-10A) in vitro. As shown in Table 4, all selected compounds showed good anticancer activities (IC50: 0.57–16.0 μM) but no significant toxicity to normal cells (IC50 > 50 μM). Additionally, compound 9h exhibited the most excellent anticancer activity with IC50 values of range from 0.57 ± 0.02 µM to 1.73 ± 0.09 µM. Considering that compound 9h exhibited excellent inhibition on HIV-1 transcription and had similar anticancer activity to the positive compound FLP, we then selected compound 9h as a candidate compound for the following biological activities study. The values are the mean ± standard deviation of triplicate measurements.
Compound
IC50 (μM)±S.D.
Cancer cells
Normal cells
A375
A549
HepG2
MCF-7
Hacat
MCF-10A
9f
13.6 ± 0.20
4.5 ± 0.13
3.25 ± 0.11
5.84 ± 0.23
>50
>50
9h
0.58±0.02
1.73±0.09
0.57±0.02
1.14±0.06
>50
>50
9j
4.10 ± 0.02
8.90 ± 0.13
16.0 ± 0.23
5.90 ± 0.13
>50
>50
9l
13.1 ± 0.20
6.1 ± 0.10
8.2 ± 0.14
7.0 ± 0.23
>50
>50
10h
10.6 ± 0.29
4.5 ± 0.13
3.25 ± 0.14
7.78 ± 0.23
>50
>50
10j
11.2 ± 0.11
7.5 ± 0.10
8.20 ± 0.31
8.80 ± 0.20
>50
>50
10l
6.6 ± 0.12
8.0 ± 0.19
10.25 ± 0.44
7.68 ± 0.11
>50
>50
FLP
0.15 ± 0.01
0.28 ± 0.03
0.81 ± 0.03
0.21 ± 0.07
>50
>50
12i
0.10 ± 0.02
0.53 ± 0.09
0.07 ± 0.04
0.10 ± 0.06
>50
>50
2.4 The inhibitory activity of compound 9 h on the HIV-1 transcription and the phosphorylation of Pol II CTD Ser2
To confirm whether compound 9h was an excellent CDK9 inhibitor, we assessed its HIV-1 transcription inhibition in a time-dependent reporter gene assay and checked the phosphorylated Ser2 (p-Ser2) level of RNAPII CTD in Hela cells treated with/without 9h by using western blot analysis. As shown in Fig. 4A, compound 9h at a lower concentration (2.0 µM) showed a time-dependent effect on the inhibition of the HIV-1 transactivation. The inhibition rate of 9h on HIV transcription reached 87% after 8 h treatment. Besides, Fig. 4B and Fig. S1 showed that 9h decreased the p-Ser2 level in a dose-dependent manner and 9h showed equal inhibitory activity as FLP at 2.0 µM. Taken together, these data indicated that compound 9h could be a potent CDK9 inhibitor working on blocking phosphorylation and repressing HIV-1 transcription.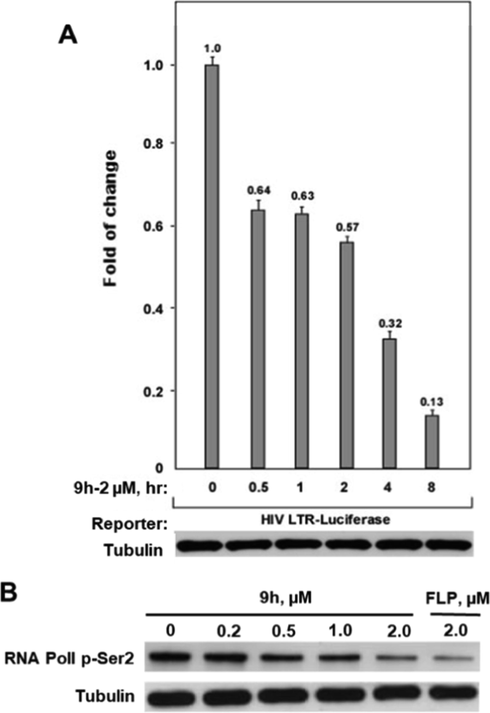
(A) The HIV-1transcription inhibition of compound 9h at different concentrations in NH2 cells. (B) Western blotting analysis of the phosphorylated Ser2(p-Ser2) level of RNAPII CTD in Hela cells treated with 9h at different concentrations.
2.5 Cell apoptosis induction of compound 9 h
Most anticancer agents suppress tumor development by inducing cell apoptosis. Moreover, previous studies have demonstrated that CDK9 inhibitors could effectively induce cell apoptosis of cancer cells. Therefore, we evaluated the ability of compound 9h to induce cell apoptosis by detecting cleaved PARP expressing levels and the percentage of apoptosis cells in 9h-treated cancer cells. First, we evaluated the effect of 9h on the induction of PARP cleavage in four cancer cell lines. As shown in Fig. 5A and Fig. S2, 9h at a concentration of >2 μM significantly induced cleaved PARP expression. In A549 cells and HepG2 cells, the PARP cleavage could be detected at 1.0 µM of 9h treatment. These results indicated that 9h induced cell apoptosis in cancer cells. Interestingly, 9h-treatment at indicated concentrations resulted in the different expression trends of full-length PARP in A375/MCF7 and HepG2 cells, which is similar to the results of 12i-treatment (Hu et al., 2020). Considering that PARP is a critical enzyme involved in DNA repair and many other biological processes, we infer that 9h may participate in various cellular processes, and then have different effects on the protein expressions of full-length PARP in different cell lines.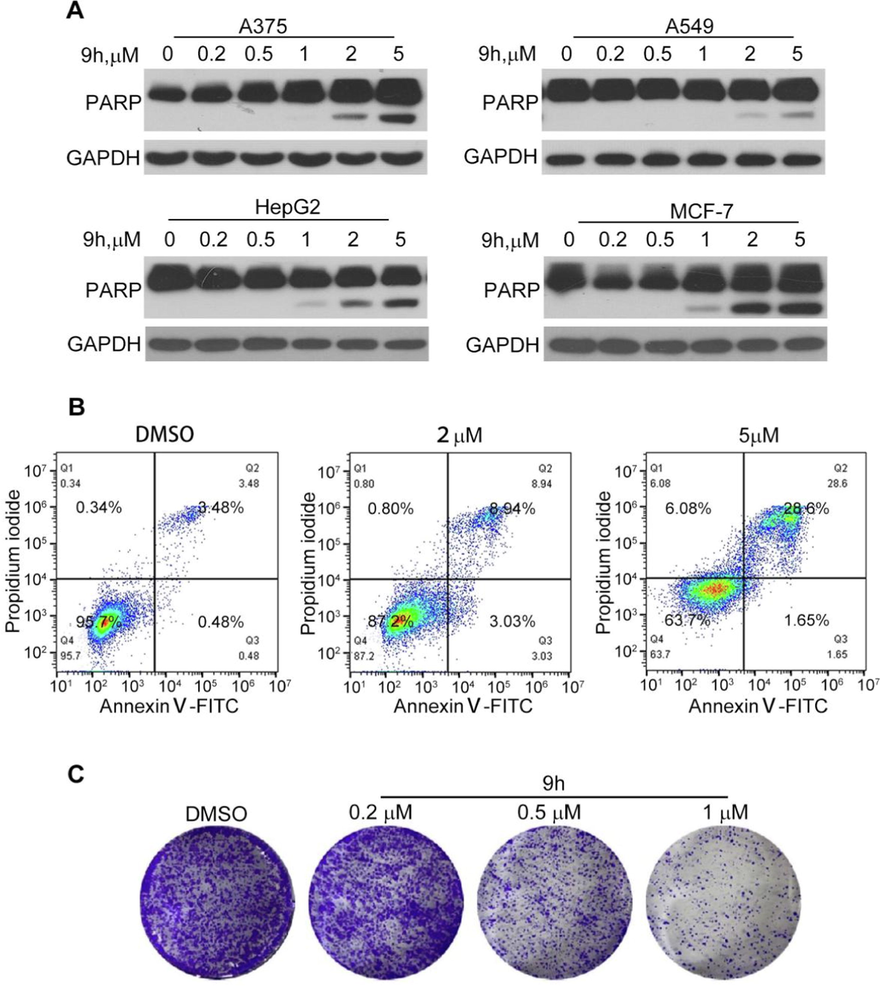
The apoptosis induction of 9h in cancer cells. (A) Induction of PARP cleavage by 9h. Different cancer cell lines were treated with 9h at indicated concentrations for 24 h. Cleaved PARP was tested by western blotting. (B) Apoptosis assay of compound 9h on HepG2 cells. HepG2 cells were treated with compound 9h at indicated concentrations for 24 h, then subjected to double stating and detected through FACS. (C) Colony formation assay of 9 h on HepG2 cells. Cells were treated with compound 9 h at indicated concentrations for 7 days, then subjected to crystal violet staining.
As compound 9h performed potent anti-proliferation against HepG2 cells (IC50 = 0.07 µM) and significant induction of cleaved PARP in HepG2 cells. To further confirm whether compound 9h could induce apoptosis, HepG2 cells were cultured in a medium containing different concentrations of compound 9h (2, 5 μM) for 24 h and then analyzed by flow cytometry after Annexin V-FITC/PI double staining. As observed in Fig. 5B and Fig. S3, the total number of apoptotic cells grew with increasing consistency. The treatment of 9h at 5 µM led to 28.6% apoptosis cells. The colony formation assay is often used to detect the cancer cell survival and proliferation abilities. Thus, we carried out a colony formation assay to detect the antitumor roles of the compound 9h. As shown in Fig. 5C, we found that compound 9h could both eliminate the colonies and decrease the colony sizes, indicating that 9h had a strong anti-survival effect on liver cancer cells and also played antiproliferation roles. These results further demonstrate that compound 9h has potent anticancer properties.
2.6 Molecular docking study
The molecular docking method is generally performed to comprehensively understand the protein-small molecule interactions at the atomic level. So, we first performed molecular docking studies to identify the orientation, binding pattern, and the interacting amino acids of 9h in CDK9 using the Induced Fit docking protocol of Schrödinger Suits (a professional drug design software). The native ligand FLP and our previously reported molecule 12i were also subjected to docking studies for comparison. Based on the docking score and docking pose, the preferential docking patterns of 9h, FLP, and 12i were produced. As shown in Fig. 6, compound 9h could dock well into the binding cavity of CDK9 and had a similar binding orientation to the native ligand FLP (Fig. S4). However, the indole scaffold of 12i made the molecular size too bulky and further increased its spatial resistance, resulting in its binding conformation opposite to that of 9h and FLP. The docking score of 9h (−7.072) was observed to be higher than that of 12i (−6.878) but lower than that of FLP (−8.442). Subsequently, we used the MMGBSA module to calculate the binding free energy of the simulated complexes 9h/CDK9, FLP/CDK9, and 12i/ CDK9. It exposed the binding free energy (BFE) of 9h (−87.12 kcal/mol) was lower than those of FLP (−68.67 kcal/mol) and 12i (−69.43 kcal/mol).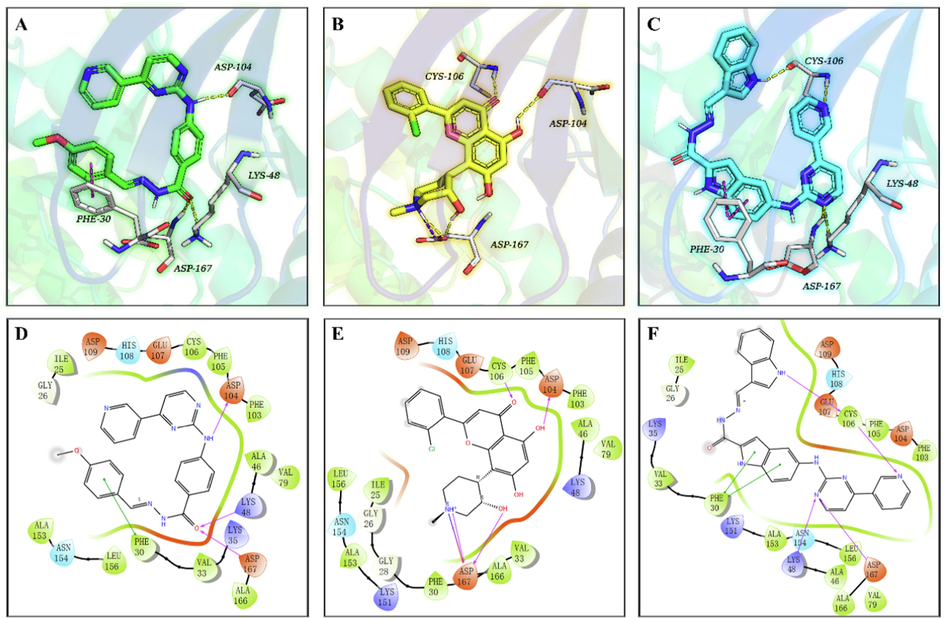
The co-crystal structure of CDK9 in complex with FLP (PDB ID: 3BLR) and proposed CDK9 binding models of 9h and 12i. Compounds 9h (A, green), FLP (B, yellow), and 12i (C, cyan) are shown in the sticks model, the protein is displayed as a colorful cartoon, and the key residues are represented as white sticks. H-bonds are indicated with yellow dashed lines, and π-π stacking interactions are shown as magenta dashes. Two-dimensional binding pattern of 9h (D), FLP (E), 12i (F) and CDK9 produced by Schrödinger. Purple solid arrows indicate hydrogen bonds, green lines indicate π-π interactions, green bands around ligands indicate hydrophobic interactions between 9h, FLP, 12i and the binding pocket residues.
Next, we analyzed the binding models of complexes 9h/CDK9, FLP/CDK9, and 12i/ CDK9. For complex 9h/CDK9, as depicted in Fig. 6A and D, the amino group on C4-position of benzene ring, serving as a hydrogen bond donor, forms a powerful hydrogen bond (2.0 Å) with residues ASP104 in the hinge region of CDK9. Oxygen atom on the carbonyl group of 9h, as a hydrogen bond acceptor, forms two hydrogen bonds to residues LYS48 and ASP167 with distances of 2.3 Å and 2.2 Å, respectively. Besides, the benzene ring of 9h forms a strong pi-pi interaction with residues PHE30 with a short π-π stacking distance of 3.3 Å. Additionally, the aromatic rings of 9h also form strong hydrophobic interactions with amino acid residues in the protein’s active site. In comparison of the 3D docked poses of 9h with those of the reference ligands (FLP and 12i), ASP167 is found to be a common interaction residue (Fig. 6). Besides, both 9h and FLP can interact with the residue ASP104, whereas 9h and 12i possess an additional common interaction with PHE30, which is not found in the native ligand FLP. However, compared with FLP and 12i, 9h lacks hydrogen bond interactions formed with CYS106 in the hinge region.
The modeling study demonstrates that 9h can competently bind to the active pocket of CDK9. Moreover, the docking results indicates that 9h (docking Score, −7.072; BFE value, −87.12 kcal/mol) has a similar or even better binding affinity with CDK9 compared to 12i (docking Score, −6.878; BFE value, −69.43 kcal/mol), which is not consistent with their inhibition effect at low concentrations on HIV-1 transcription. Thus, we speculate that the physicochemical properties (e.g. the low membrane permeability) of 9h might play a critical role in its poor absorption, resulting in its low inhibition of CDK9 activity at low concentrations (0.2 μM and 2.0 μM).
3 Conclusion
In summary, we described the continuous optimization of previously disclosed (5-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)-1H-indole carbohydrazide derivatives as CDK9 inhibitors. The strategy was to replace the indole ring with a phenyl ring, change the link position of pyridine at the 4-position of pyrimidine, and introduced different substitutions at the position of the methylene group, producing three series of benzene carbohydrazide derivatives. Finally, 42 new benzene carbohydrazide derivatives bearing a (pyridyl pyrimidin-2-yl)amino moiety on the para-position were synthesized and evaluated for their inhibition effect on HIV-1 transcription. We then discovered that almost all synthetic compounds at 20 μM showed excellent inhibition of HIV-1 transcription, implying they were potent CDK9 inhibitors. In particular, compound 9h, which displayed excellent anti-proliferative potency in cancer cells but not in normal cells, could suppress both HIV-1 transcription and the phosphorylation at Serine2 of the RNAPIICTD in a dose-dependent manner. Meanwhile, compound 9h was found to exert its anti-proliferative effect by inducing apoptosis in cancer cells. Finally, the molecular docking analysis showed the mode of action of compound 9h with CDK9 and further proved its CDK9 inhibitory. Therefore, our study presents a novel series of benzene carbohydrazide derivatives as CDK9 inhibitors, and compound 9h represents a new scaffold to target CDK9 for novel antitumor drug discovery.
4 Experimental section
4.1 Chemistry
All reagents were purchased and used without further purification unless otherwise indicated. Reactions were magnetically stirred and monitored by thin-layer chromatography (TLC) on Merck silica gel 60F-254 by fluorescence. All of the final compounds were purified by column chromatography. 1H NMR and 13C NMR spectra were obtained using a Bruker AV2 600 Ultrashield spectrometer at 600 and 150 MHz, and chemical shifts were given in parts per million (ppm) relative to tetramethylsilane (TMS) as an internal standard. Multiplicities were abbreviated as follows: single (s), doublet (d), doublet–doublet (dd), doublet-triplet (dt), triplet (t), triplet–triplet (tt), triplet-doublet (td), quartet (q), quartet-doublet (qd), multiplet (m), and broad signal (brs). High-resolution mass spectral (HRMS) data were acquired on a Q Exactive. Melting points were measured on SGW X-4 micro-melting point spectrometer and were uncorrected.
4.1.1 Ethyl 4-(4-(pyridin-3-yl)pyrimidin-2-ylamino)benzoate(3)
A mixture of 2.74 g (16.6 mmol) of ethyl 4-aminobenzoate (1), 1.59 g (38.0 mmol) cyanamide, in 100 mL of ethanol (EtOH). While being stirred concentrated HCl (2.1 mL, 24.84 mmol) was dropped slowly into the mixture. The mixture was heated to reflux for 24 h. EtOH was removed by reduced pressure distillation, and water (30 mL) was added to the residue. Under 0 °C, NH4NO3 (2.64 g, 33.12 mmol) solution was dropped during 30 min. Then the mixture was stirred for 60 min, and filtered, giving the crude product. The crude products were purified by recrystallization with ether and dried to afford ethyl 4-guanidinobenzoate (2). After the ethyl 4-guanidinobenzoate (2) was in to the round-bottom flask, 3-dimethylamino-1-(3-pyridyl)-2-propen-1-one (1.76 g, 10 mmol), sodium hydroxide (0.48 g,12 mmol) was dissolved in EtOH (20 mL). The mixture was stirred and heated to reflux for 48 h. when TLC indicated that the reaction had gone to completion. On cooling, Precipitated complex was filtered, washed with ethyl acetate and dried to afford ethyl 4-(4-(pyridin-3-yl)pyrimidin-2-ylamino)benzoate (3) as a yellow solid (Pan et al., 2013). Yield: 3.2 g (60.2%),1H NMR(600 MHz, DMSO‑d6): δ 10.23 (s, 1H), 9.37 (d, J = 2.0 Hz, 1H), 8.75 (dd, J = 1.5, 4.8 Hz, 1H), 8.69 (d, J = 5.1 Hz, 1H), 8.54(td, J = 1.8, 8.0 Hz, 1H), 7.97–8.01 (m, 2H), 7.93–7.95 (m, 2H), 7.62 (d, J = 5.1 Hz, 1H), 7.61 (d, J = 5.0 Hz, 1H), 4.29 (q, J = 7.0 Hz, 2H), 1.32 (t, J = 7.0 Hz, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 166.0, 162.3, 160.2, 160.0, 152.1, 148.7, 145.5, 135.0, 132.5, 130.6,124.5, 122.7,118.3, 109.7, 60.7, 14.7; HRMS (+): calcd for C18H16N4O2 [M + H]+ 321.1346, found 321.1344; calcd for C18H16N4O2Na [M + Na]+ 343.1165, found 343.1165.
Compounds 4–5 were prepared by the methods described previously (Pan et al., 2013).
ethyl 4-(4-(pyridin-2-yl)pyrimidin-2-ylamino)benzoate(4)yellow solid. Yield: (64.2%);1H NMR (600 MHz, CDCl3): δ 8.73 (d, J = 4.4 Hz, 1H), 8.62 (d, J = 5.0 Hz, 1H), 8.42 (d, J = 7.7 Hz, 1H), 8.07 (d, J = 8.2 Hz, 2H), 7.89–7.91 (m, 1H), 7.87 (d, J = 5.7 Hz, 1H), 7.81 (d, J = 8.0 Hz, 2H), 7.56 (brs, 1H), 7.40–7.45 (m, 1H), 4.38 (q, J = 7.0 Hz, 2H), 1.41 (t, J = 7.0 Hz, 3H).
ethyl 4-(4-(pyridin-4-yl)pyrimidin-2-ylamino)benzoate(5)yellow solid. Yield: (68.4%); 1H NMR (600 MHz, DMSO‑d6): δ 10.28 (s, 1H), 8.79–8.83 (m, 2H), 8.74 (d, J = 5.13 Hz, 1H), 8.10–8.14 (m, 2H), 7.98–8.01 (m, 2H), 7.94–7.96 (m, 2H), 7.65 (d, J = 5.14 Hz, 1H), 4.29 (q, J = 7.09 Hz, 2H), 1.32 (t, J = 7.15 Hz, 3H).
4.1.2 4-(4-(pyridin-3-yl)pyrimidin-2-ylamino)benzohydrazide (6)
To a solution of ethyl 4-(4-(pyridin-3-yl)pyrimidin-2-ylamino)benzoate3 (3.2 g,10 mmol) in 95% EtOH (10 mL) hydrazine hydrate (10 mL) was added and the reaction mixture was heated under reflux for 8 h. The hydrazide was obtained in quantitative yields directly after evaporation of the reaction solution without any additional purification step (Nayyar et al., 2007). White solid. Yield: 87.7%. 1H NMR (600 MHz, DMSO‑d6): δ 10.05 (s, 1H), 9.61 (s, 1H), 9.36 (d, J = 1.8 Hz, 1H), 8.75 (dd, J = 1.5, 4.8 Hz, 1H), 8.67 (d, J = 5.0 Hz, 1H), 8.52 (td, J = 2.0, 8.0 Hz, 1H), 7.91 (d, J = 8.8 Hz, 2H), 7.82 (d, J = 8.8 Hz, 2H), 7.61 (dd, J = 4.9, 7.8 Hz, 1H), 7.58 (d, J = 5.0 Hz, 1H), 4.45 (br s, 1H); 13C NMR (150 MHz, DMSO‑d6): δ 166.2, 162.2, 160.4, 159.9, 152.1, 148.7, 143.5, 134.9, 132.6, 128.1, 126.4, 124.5, 118.3, 109.4; HRMS (+): calcd for C16H14N6O [M + H]+ 307.1302 found 307.1302, calcd for 315.0852 C16H14N6ONa [M + Na]+ 329.1121 found 329.1120.
Compounds 7–8 were prepared by the methods described previously (Pan et al., 2013).
4-(4-(pyridin-2-yl)pyrimidin-2-ylamino)benzohydrazide (7). White solid. Yield: 83.3%; 1H NMR (600 MHz, DMSO‑d6): δ 10.06 (brs, 1H), 9.62 (brs, 1H), 8.77 (d, J = 3.0 Hz, 1H), 8.70 (d, J = 4.8 Hz, 1H), 8.43 (d, J = 7.5 Hz, 1H), 8.07 (t, J = 7.4 Hz, 1H), 7.93 (d, J = 8.4 Hz, 2H), 7.84 (d, J = 8.2 Hz, 2H), 7.79 (d, J = 4.8 Hz, 1H), 7.64–7.54 (m, 1H), 4.50 (brs, 2H); 13C NMR (150 MHz, DMSO‑d6): δ 166.3, 163.4, 160.3, 160.1, 154.0, 150.2, 143.6, 138.1, 128.2, 126.3, 121.6, 118.3, 109.1; HRMS: calcd for C16H14N6O [M + H]+ 307.1302 found 307.1300, calcd for C16H14N6ONa [M + Na]+ 329.1121 found 329.1118.
4-(4-(pyridin-4-yl)pyrimidin-2-ylamino)benzohydrazide (8). White solid. Yield:86%; 1H NMR (600 MHz, DMSO‑d6):δ 10.11 (s, 1H), 9.62 (s, 1H), 8.86–8.76 (m, 2H), 8.71 (d, J = 5.1 Hz, 1H), 8.13–8.06 (m, 2H), 7.91 (d, J = 8.8 Hz, 2H), 7.83 (d, J = 8.8 Hz, 2H), 7.60 (s, 1H), 4.44 (brs, 2H);13C NMR (150 MHz, DMSO‑d6): δ 166.2, 162.0, 160.5, 160.4, 151.1, 144.2, 143.4, 130.7, 128.2, 126.5, 121.4, 118.4, 109.7; HRMS: calcd for C16H14N6O [M + H]+ 307.1302 found 307.1299, calcd for C16H14N6ONa [M + Na]+ 329.1121 found 329.1119.
4.1.3 N-Substituted Methyl subunit-4-((4-Pyridine-3-yl)Pyrimidine-2-yl)Amino Benzoyl hydrazide derivatives (9a-9n)
To a solution of 4-(4-(pyridin-3-yl)pyrimidin-2-ylamino)benzohy drazide (0.075 g, 0.25 mmol) in ethanol (10 mL), Substituted aldehyde (0.3 mmol) was added, and the reaction mixture was heated to reflux for 8 h, and monitored by TLC analysis. After completion of the reaction, the mixture was concentrated. Then the residue was purified by column chromatography to get yellow solid with the yield of 80–92%.
9a: Yield: 89%; Rf, 0.23(CH3OH/CH2Cl2 = 1:50); 1H NMR (600 MHz, DMSO‑d6): δ 11.32 (brs, 1H), 10.22 (s, 1H), 8.91 (d, J = 6.2 Hz, 2H), 8.78 (d, J = 5.1 Hz, 1H), 8.31 (d, J = 6.2 Hz, 2H), 7.95 (d, J = 8.6 Hz, 2H), 7.88 (d, J = 8.6 Hz, 2H), 7.76 (brs, 1H), 7.69 (d, J = 5.1 Hz, 1H), 2.34–2.21(m, 2H), 1.07 (t, J = 7.4 Hz, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 166.3, 160.3, 160.1, 147.7, 147.5, 145.2, 136.4, 129.1, 128.8, 125.0, 125.0, 118.5, 118.4, 109.9, 109.5, 25.9, 11.2; HRMS (+): calcd for C19H19N6O [M + H]+ 347.1615, found 347.1616, calcd for C19H18N6ONa [M + Na]+ 369.1434, found 369.1435.
9b: Yield: 86%; Rf, 0.26(CH3OH/CH2Cl2 = 1:50); 1H NMR (600 MHz, DMSO‑d6): δ 11.30 (brs, 1H), 10.14 (s, 1H), 9.41 (d, J = 1.6 Hz, 1H), 8.82 (dd, J = 1.5, 4.9 Hz, 1H), 8.70 (d, J = 5.1 Hz, 1H), 8.64–8.69 (m, 1H), 7.95 (d, J = 8.4 Hz, 2H), 7.87 (d, J = 8.6 Hz, 2H), 7.74 (dd, J = 5.0, 7.6 Hz, 2H), 7.63 (d, J = 5.1 Hz, 1H), 2.25 (q, J = 6.8 Hz, 2H), 1.53 (sxt, J = 7.3 Hz, 2H), 0.94 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 166.5, 166.3, 161.6, 160.3, 160.1, 151.8, 150.7, 147.4, 136.5, 133.1, 129.1, 128.8, 125.0, 118.5, 118.4, 109.5, 34.4, 20.1, 20.0, 14.1; HRMS (+): calcd for C20H21N6O [M + H]+ 361.1771, found 361.1768, calcd for C20H20N6ONa [M + Na]+ 383.1591, found 383.1586.
9c: Yield: 87%; Rf, 0.31 (CH3OH/CH2Cl2 = 1:20); 1H NMR (600 MHz, DMSO‑d6): δ 11.84 (brs, 1H), 10.28 (s, 1H), 9.52 (d, J = 1.8 Hz, 1H), 8.93 (dd, J = 1.5, 4.9 Hz, 1H), 8.81 (d, J = 5.1 Hz, 1H), 8.80–8.78(m, 1H), 8.57 (brs, 1H), 8.11–8.07 (m, 2H), 8.01–8.06 (m, 2H), 7.85 (dd, J = 5.2, 8.16 Hz, 1H), 7.83 (d, J = 7.0 Hz, 1H), 7.73 (d, J = 5.1 Hz, 1H), 7.50–7.60 (m, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.2, 161.3, 160.3, 160.2, 149.8, 147.5, 146.6, 144.1, 137.5, 135.0, 133.5, 130.4, 129.3, 129.0, 127.5, 126.3, 125.4, 118.4, 109.6; HRMS (+): calcd for C23H19N6O[M + H]+ 395.1615, found 395.1613, calcd for C23H18N6ONa [M + Na]+ 417.1434, found 417.1432.
9d: Yield: 90%; Rf, 0.53(CH3OH/CH2Cl2 = 1:20); 1H NMR (600 MHz, DMSO‑d6): δ 11.68 (brs, 1H), 10.16 (s, 1H), 9.39 (d, J = 1.5 Hz, 1H), 8.78 (dd, J = 1.5, 4.6 Hz, 1H), 8.72–8.68 (m, 1H), 8.58 (d, J = 7.9 Hz, 1H), 8.43 (brs, 1H), 8.02–7.98 (m, 2H), 7.96–7.92 (m, 2H), 7.66–7.60 (m, 5H), 7.28 (d, J = 7.5 Hz, 2H), 2.35 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.1, 162.0, 160.4, 160.1, 151.6, 148.2, 135.5, 132.8, 132.3, 130.0, 129.9, 129.1, 129.0, 128.8, 127.5, 124.7, 118.4, 118.4, 109.5, 21.5; HRMS (+): calcd for C24H21N6O [M + H]+ 409.1771, found 409.1772, calcd for C24H20N6ONa [M + Na]+ 431.1591, found 431.1592.
9e: Yield: 90%; Rf, 0.61(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.89 (brs, 1H), 10.21 (s, 1H), 9.44 (d, J = 1.5 Hz, 1H), 8.85 (dd, J = 1.0, 4.9 Hz, 1H), 8.77–8.69 (m, 3H), 8.03–7.98 (m, 2H), 7.98–7.94 (m, 2H), 7.79 (dd, J = 4.9, 7.9 Hz, 1H), 7.65 (d, J = 5.1 Hz, 1H), 7.50 (q, J = 6.4 Hz, 1H), 7.34–7.29 (m, 2H); 13C NMR (150 MHz, DMSO‑d6): δ 162.6 (d, J = 166.1 Hz),161.2, 160.4, 160.3, 160.3, 149.8, 146.6, 144.2, 140.1, 137.6, 133.5, 132.3 (d, J = 8.8 Hz), 129.1, 126.7, 126.1, 125.5, 125.4 (d, J = 3.3 Hz), 122.5, 122.5, 118.5, 116.5 (d, J = 20.9 Hz), 109.7; HRMS (+): calcd for C23H18FN6O [M + H]+ 413.1521, found 413.1516, calcd for C23H17FN6ONa [M + Na]+ 436.1340, found 436.1366.
9f: Yield: 89%; Rf, 0.46(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.55 (brs, 1H), 10.17 (s, 1H), 9.42 (d, J = 1.8 Hz, 1H), 8.83 (dd, J = 1.5, 4.9 Hz, 1H), 8.72–8.69 (m, 2H), 8.36 (s, 1H), 8.00–7.96 (m, 2H), 7.94–7.90 (m, 2H), 7.76 (dd, J = 4.9, 8.0 Hz, 1H), 7.63 (d, J = 5.1 Hz, 1H), 7.57 (d, J = 8.1 Hz, 2H), 6.85 (d, J = 8.4 Hz, 2H); 13C NMR (150 MHz, DMSO‑d6): δ 163.0, 161.2, 160.3, 160.2, 159.8, 159.0, 158.8, 149.8, 148.0, 146.6, 143.9, 137.5, 133.5, 129.2, 128.9, 126.5, 125.9, 125.4, 118.4, 116.2, 109.6; HRMS (+): calcd for C23H19N6O2 [M + H]+ 411.1564, found 411.1563, calcd for C23H18N6O2Na [M + Na]+ 433.1383, found 433.1383.
9g: Yield: 85%; Rf, 0.11(CH3OH/CH2Cl2 = 1:5); 1H NMR (600 MHz, DMSO‑d6): δ 11.90 (brs, 1H), 10.17 (s, 1H), 9.38 (d, J = 1.8 Hz, 1H), 8.76 (dd, J = 1.5, 4.6 Hz, 1H), 8.69 (d, J = 5.1 Hz, 1H), 8.55 (td, J = 2.0 8.0 Hz, 1H), 8.53–8.50 (m, 1H), 8.01 (t, J = 8.5 Hz, 4H), 7.98–7.94 (m, 2H), 7.85 (d, J = 7.1 Hz, 2H), 7.63 (d, J = 5.1 Hz, 1H), 7.61 (d, J = 5.1 Hz, 2H); 13C NMR (150 MHz, DMSO‑d6): δ 167.4, 162.2, 160.3, 160.3, 160.0, 152.1, 148.7, 146.3, 139.0, 135.0, 132.5, 132.0, 130.3, 129.1, 128.3, 127.5, 125.9, 124.5, 118.4, 109.6; HRMS (−): calcd for C24H17N6O3 [M−H]- 437.1938 found 437.1940.
9h: Yield: 82%; Rf, 0.57(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.62 (s, 1H), 10.17 (s, 1H), 9.41 (d, J = 1.6 Hz, 1H), 8.80 (dd, J = 1.6, 4.9 Hz, 1H), 8.70 (d, J = 5.1 Hz, 1H), 8.63 (td, J = 1.7, 8.0 Hz, 1H), 8.41 (s, 1H), 8.01–7.97 (m, 2H), 7.95–7.91 (m, 2H), 7.72–7.69 (m, 1H), 7.69–7.66 (m, 2H), 7.62 (d, J = 5.1 Hz, 1H), 7.03 (d, J = 8.6 Hz, 2H), 3.82 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.0, 161.8, 161.2, 160.4, 160.1, 151.2, 147.9, 147.4, 144.1, 136.0, 132.9, 129.1, 128.9, 127.5, 126.4, 124.9, 118.4, 114.8, 109.6, 55.8; HRMS (+): calcd for C24H21N6O2 [M + H]+ 425.1721, found 425.1717, calcd for C24H20N6O2Na [M + Na]+ 447.154 found 447.1539.
9i: Yield:84%; Rf, 0.51 (CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.78 (brs, 1H), 10.18 (s, 1H), 9.42 (d, J = 1.6 Hz, 1H), 8.81 (dd, J = 1.5, 4.9 Hz, 1H), 8.75 (s, 1H), 8.73–8.68 (m, 1H), 8.65 (td, J = 1.8, 8.1 Hz, 1H), 8.02–7.94 (m, 4H), 7.71 (dd, J = 4.9, 7.9 Hz, 1H), 7.63 (d, J = 5.1 Hz, 1H), 7.49 (d, J = 7.1 Hz, 1H), 7.18–7.09 (m, 2H), 3.85 (s, 3H), 3.81 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 161.6, 160.3, 160.2, 158.7, 153.1, 150.7, 148.4, 147.4, 143.1, 136.5, 133.1, 129.2, 129.0, 128.4, 125.1, 124.8, 118.5, 118.4, 117.4, 114.5, 109.6, 61.7, 56.2; HRMS (+): calcd for C25H23N6O3 [M + H]+ 455.1826, found 455.1820, calcd for C25H22N6O3Na [M + Na]+ 477.1646, found 477.1641.
9j: Yield: 85.6%; Rf, 0.56(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.62 (s, 1H), 10.15 (s, 1H), 9.39 (d, J = 1.8 Hz, 1H), 8.76 (dd, J = 1.4, 4.7 Hz, 1H), 8.69 (d, J = 5.1 Hz, 1H), 8.55 (td, J = 1.9, 7.9 Hz, 1H), 8.40 (s, 1H), 8.02–7.98 (m, 2H), 7.95–7.91 (m, 2H), 7.64–7.62 (m, 1H), 7.61 (d, J = 5.1 Hz, 1H), 7.35 (s, 1H), 7.21 (d, J = 7.9 Hz, 1H), 7.04 (d, J = 8.4 Hz, 1H), 3.84 (s, 3H), 3.82 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.0, 162.2, 160.4, 160.0, 152.1, 151.1, 149.5, 148.6, 147.8, 144.1, 135.0, 132.6, 128.9, 127.7, 126.4, 124.5, 122.2, 118.4, 112.0, 109.5, 108.6, 56.0, 55.9; HRMS (+): calcd for C25H23N6O3 [M + H]+ 455.1826, found 455.1825, calcd for C25H22N6O3Na [M + Na]+ 477.1646, found 477.1646.
9k: Yield: 87%; Rf, 0.76(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.84 (brs, 1H), 10.20 (s, 1H), 9.44 (d, J = 1.8 Hz, 1H), 9.18 (s, 1H), 8.84 (dd, J = 1.4, 4.9 Hz, 1H), 8.73 (d, J = 5.1 Hz, 1H), 8.71 (td, J = 1.7, 8.1 Hz, 1H), 8.06 (d, J = 9.0 Hz, 1H), 8.02 (s, 4H), 7.92 (d, J = 8.1 Hz, 1H), 7.76 (dd, J = 5.0, 8.0 Hz, 1H), 7.64 (d, J = 5.1 Hz, 1H), 7.60 (t, J = 7.4 Hz, 1H), 7.54 (d, J = 9.0 Hz, 1H), 7.44 (t, J = 7.2 Hz, 1H), 4.03 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.1, 161.2, 160.3, 160.2, 158.8, 158.2, 149.7, 146.5, 145.2, 144.1, 137.6, 133.6, 133.0, 131.2, 129.2, 129.1, 128.9, 128.4, 126.5, 126.4, 125.5, 124.5, 118.5, 114.9, 113.9, 109.6, 57.1; HRMS (+): calcd for C28H23N6O2 [M + H]+ 475.1877, found 475.1879, calcd for C28H22N6O2Na [M + Na]+ 497.1696, found 497.1698.
9l: Yield: 88%; Rf, 0.87(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.70 (brs, 1H), 10.19 (s, 1H), 9.43 (d, J = 1.8 Hz, 1H), 8.84 (dd, J = 1.5, 4.9 Hz, 1H), 8.72 (d, J = 5.1 Hz, 1H), 8.70 (s, 1H), 8.36 (brs, 1H), 8.01–7.97 (m, 2H), 7.92 (d, J = 8.8 Hz, 2H), 7.86 (s, 1H), 7.76 (dd, J = 5.0, 8.0 Hz, 1H), 7.64 (d, J = 5.1 Hz, 1H), 6.93 (d, J = 2.6 Hz, 1H), 6.65 (d, J = 1.3 Hz, 1H); 13C NMR (150 MHz, DMSO‑d6): δ 161.2, 160.3, 160.2, 159.0, 150.1, 149.8, 146.6, 145.5, 144.1, 137.5, 137.4, 133.5, 129.0, 126.2, 125.4, 118.4, 113.6, 112.6, 109.7; HRMS (+): calcd for C21H17N6O2 [M + H]+ 385.1408, found 385.1402, calcd for C21H16N6O2Na [M + Na]+ 407.1227, found 407.1223.
9m: Yield: 86%; Rf, 0.79(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.70 (brs, 1H), 10.19 (s, 1H), 9.43 (d, J = 1.6 Hz, 1H), 8.90–8.81 (m, 1H), 8.73–8.68(m, 3H), 7.99 (d, J = 8.4 Hz, 2H), 7.92 (d, J = 8.6 Hz, 2H), 7.77 (dd, J = 4.9, 7.9 Hz, 1H), 7.67 (d, J = 4.6 Hz, 1H), 7.64 (d, J = 5.1 Hz, 1H), 7.47 (d, J = 2.7 Hz, 1H), 7.18–7.12 (m, 1H); 13C NMR (150 MHz, DMSO‑d6): δ 163.0, 161.5, 160.3, 160.2, 150.3, 147.1, 144.1, 142.7, 139.8, 136.9, 133.3, 131.1, 129.2, 128.9, 128.3, 126.3, 125.2, 118.4, 109.6; HRMS (+): calcd for C21H17N6OS [M + H]+ 401.1179, found 401.1176, calcd for C21H16N6OSNa [M + Na]+ 423.0999, found 423.0996.
9n: Yield: 87%; Rf, 0.86(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.99 (brs, 1H), 10.20 (s, 1H), 9.40 (d, J = 1.6 Hz, 1H), 8.79 (dd, J = 1.5, 4.8 Hz, 1H), 8.70 (d, J = 5.1 Hz, 1H), 8.64 (d, J = 4.8 Hz, 1H), 8.60 (td, J = 1.7, 8.0 Hz, 1H), 8.50 (brs, 1H), 8.03–8.00 (m, 2H), 7.98–7.95 (m, 2H), 7.95–7.90 (m, 1H), 7.67 (dd, J = 4.9, 7.9 Hz, 1H), 7.63 (d, J = 5.1 Hz, 1H), 7.48–7.42(m, 1H); 13C NMR (150 MHz, DMSO‑d6): δ 161.9, 160.3, 160.1, 153.5, 151.4, 149.6, 148.0, 147.2, 144.4, 137.9, 135.8, 132.8, 129.2, 125.9, 124.9, 124.8, 122.3, 120.6, 118.4, 109.6; HRMS (+): calcd for C22H18N7O [M + H]+ 396.1567, found 396.1568, calcd for C22H17N7ONa [M + Na]+ 418.1387, found 418.1386.
4.1.4 N-Substituted methyl subunit-4-((4-Pyridine-2-yl)Pyrimidine-2-yl)Amino Benzoyl hydrazide derivatives (10a-10n) and N-Substituted methyl subunit-4-((4-Pyridine-4-yl)Pyrimidine-2-yl)Amino Benzoyl hydrazide derivatives (11a-11n)
Compounds 10a-10n and 11a-11n were prepared by the same methods with compounds 9a-9n.
10a: Yield: 85%; Rf, 0.25(CH3OH/CH2Cl2 = 1:20); 1H NMR (600 MHz, DMSO‑d6): δ 11.30 (s, 1H), 10.11 (s, 1H), 8.79–8.76 (m, 1H), 8.71 (d, J = 5.1 Hz, 1H), 8.44 (d, J = 7.7 Hz, 1H), 8.07 (dt, J = 1.74, 7.75 Hz, 1H), 7.98 (d, J = 8.4 Hz, 2H), 7.88 (d, J = 8.6 Hz, 2H), 7.80 (d, J = 4.9 Hz, 1H), 7.77 (brs, 1H), 7.60 (ddd, J = 0.9, 4.7, 7.4 Hz, 1H), 2.33–2.25 (m, 2H), 1.07 (t, J = 7.4 Hz, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.4, 162.9, 160.3, 160.1, 153.9, 152.7, 150.2,144.1,138.1, 128.8,126.4,126.3, 121.6, 118.2, 109.2, 25.9, 11.2; HRMS (+): calcd for C19H19N6O [M + H]+ 347.1615, found 347.1615, calcd for C19H18N6ONa [M + Na]+ 369.1434, found 369.1434.
10b: Yield: 78%; Rf, 0.29(CH3OH/CH2Cl2 = 1:20); 1H NMR (600 MHz, DMSO‑d6): δ 11.30 (s, 1H), 10.11 (s, 1H), 8.80–8.76(m,1H), 8.71 (d, J = 5.1 Hz, 1H), 8.44 (d, J = 7.9 Hz, 1H), 8.07 (dt, J = 1.6, 7.7 Hz, 1H), 7.98 (d, J = 8.4 Hz, 2H), 7.88 (d, J = 8.6 Hz, 2H), 7.80 (d, J = 5.1 Hz, 1H), 7.77–7.72 (m, 1H), 7.60 (ddd, J = 1.1, 4.8, 7.5 Hz, 1H), 2.38–2.17 (m, 2H), 1.60–1.44 (m, 2H), 0.95 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.4, 162.9, 160.3, 160.1, 153.9, 151.8, 150.2, 144.1, 138.1, 128.8, 126.3, 121.6, 118.2, 109.2, 34.4, 20.0, 14.1; HRMS (+): calcd for C20H21N6O [M + H]+ 361.1771, found 361.1769, calcd for C20H20N6ONa [M + Na]+ 383.1591, found 383.1588.
10c: Yield: 85%; Rf, 0.34(CH3OH/CH2Cl2 = 1:20); 1H NMR (600 MHz, DMSO‑d6): δ 11.76 (brs, 1H), 10.16 (s, 1H), 8.78 (dd, J = 0.7, 4.58 Hz, 1H), 8.73 (d, J = 4.9 Hz, 1H), 8.48 (brs, 1H), 8.46 (d, J = 7.9 Hz, 1H), 8.08 (dt, J = 1.6, 7.7 Hz, 1H), 8.04–8.01 (m, 2H), 7.96 (d, J = 8.8 Hz, 2H), 7.82 (d, J = 4.9 Hz, 1H), 7.75 (d, J = 6.6 Hz, 2H), 7.60 (ddd, J = 1.0, 4.7, 7.5 Hz, 1H), 7.51–7.41 (m, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.4, 163.2, 160.2, 160.2, 153.9, 150.2, 147.5, 144.3, 138.1, 135.0, 130.4, 129.3, 129.0, 127.5, 126.3, 126.1, 121.6, 118.3, 109.3; HRMS (+): calcd for C23H19N6O [M + H]+ 395.1615, found 395.1615, calcd for C23H18N6ONa [M + Na]+ 417.1434, found 417.1434.
10d: Yield: 87%; Rf, 0.40 (CH3OH/CH2Cl2 = 1:20); 1H NMR (600 MHz, DMSO‑d6): δ 11.68 (brs, 1H), 10.15 (s, 1H), 8.77 (d, J = 4.0 Hz, 1H), 8.72 (d, J = 5.1 Hz, 1H), 8.50–8.41 (m, 2H), 8.07 (dt, J = 1.5, 7.7 Hz, 1H), 8.04–8.00 (m, 2H), 7.95 (d, J = 8.6 Hz, 2H), 7.81 (d, J = 5.1 Hz, 1H), 7.63 (d, J = 7.3 Hz, 2H), 7.62–7.58(m, 1H), 7.28 (d, J = 7.7 Hz, 2H), 2.35 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.4, 160.2, 160.2, 153.9, 150.2, 147.5, 138.5, 138.1, 134.9, 131.1, 129.2, 129.0, 127.7, 126.3, 126.1, 124.9, 121.6, 118.3, 109.3, 21.4; HRMS (+): calcd for C24H21N6O [M + H]+ 409.1771, found 409.1774, calcd for C24H20N6ONa [M + Na]+ 431.1591, found 431.1594.
10e: Yield: 84%; Rf, 0.53(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.89 (brs, 1H), 10.17 (s, 1H), 8.78 (d, J = 4.0 Hz, 1H), 8.73 (d, J = 4.9 Hz, 2H), 8.46 (d, J = 7.9 Hz, 1H), 8.08 (dt, J = 1.6, 7.7 Hz, 1H), 8.05–8.00 (m, 2H), 7.99–7.94 (m, 3H), 7.82 (d, J = 4.9 Hz, 1H), 7.64–7.58 (m, 1H), 7.50 (q, J = 6.3 Hz, 1H), 7.36–7.27 (m, 2H); 13C NMR (150 MHz, DMSO‑d6): δ 163.4, 163.2, 161.2(d, J = 249.8 Hz), 160.2, 160.2, 153.9, 150.2, 144.4, 140.1, 138.2, 132.3 (d, J = 8.8 Hz), 129.1, 126.7, 126.3, 125.9, 125.4 (d, J = 2.2 Hz), 122.5, 121.6, 118.3, 116.5 (d,J = 19.8 Hz), 109.3; HRMS (+): calcd for C23H18FN6O [M + H]+ 413.1521, found 413.1522, calcd for C23H17FN6ONa [M + Na]+ 435.1340,found 435.1342.
10f: Yield: 83%; Rf, 0.45(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.54 (s, 1H), 10.14 (s, 1H), 9.92 (s, 1H), 8.77 (d, J = 4.0 Hz, 1H), 8.72 (d, J = 4.9 Hz, 1H), 8.45 (d, J = 7.9 Hz, 1H), 8.37 (s, 1H), 8.08 (dt, J = 1.6, 7.7 Hz, 1H), 8.01 (d, J = 8.4 Hz, 2H), 7.94 (d, J = 8.4 Hz, 2H), 7.81 (d, J = 4.9 Hz, 1H), 7.62–7.59 (m, 1H), 7.57 (d, J = 8.2 Hz, 2H), 6.85 (d, J = 8.2 Hz, 2H); 13C NMR (150 MHz, DMSO‑d6): δ 163.4, 162.9, 160.3, 160.2, 159.8, 153.9, 150.2, 147.9, 144.1, 138.1, 129.2, 128.9, 126.4, 126.3, 126.0, 121.6, 118.3, 116.2, 109.2; HRMS (+): calcd for C23H19N6O2 [M + H]+ 411.1564, found 411.1564, calcd for C23H18N6O2Na [M + Na]+ 433.1383,found 433.1385.
10g: Yield: 80%; Rf, 0.10(CH3OH/CH2Cl2 = 1:5); 1H NMR (600 MHz, DMSO‑d6): δ 11.91 (brs, 1H), 10.17 (s, 1H), 8.78 (d, J = 4.2 Hz, 1H), 8.73 (d, J = 5.1 Hz, 1H), 8.52 (brs, 1H), 8.46 (d, J = 7.9 Hz, 1H), 8.08 (dt, J = 1.3, 7.8 Hz, 1H), 8.05–8.00 (m, 4H), 7.97 (d, J = 8.6 Hz, 2H), 7.86 (d, J = 7.5 Hz, 2H), 7.83–7.80 (m, 1H), 7.60 (dd, J = 4.9, 7.1 Hz, 1H); 13C NMR (150 MHz, DMSO‑d6): δ 167.4, 162.2, 160.3, 160.3, 160.0, 152.1, 148.7, 146.3, 139.0, 135.0, 132.5, 132.0, 130.3, 129.1, 128.3, 127.5, 125.9, 124.5, 118.4, 109.6; HRMS (−): calcd for C24H17N6O3 [M−H]- 437.1938, found 437.1940.
10h: Yield: 86%; Rf, 0.48(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.61 (s, 1H), 10.15 (s, 1H), 8.81–8.76 (m, 1H), 8.72 (d, J = 4.9 Hz, 1H), 8.45 (d, J = 7.7 Hz, 1H), 8.42 (s, 1H), 8.08 (dt, J = 1.6, 7.7 Hz, 1H), 8.03–7.99(m, 2H), 7.94 (d, J = 8.8 Hz, 2H), 7.81 (d, J = 5.1 Hz, 1H), 7.69 (d, J = 8.1 Hz, 2H), 7.60 (ddd, J = 0.9, 4.8, 7.3 Hz, 1H), 7.04 (d, J = 8.4 Hz, 2H), 3.82 (s, 3H) 13C NMR (150 MHz, DMSO‑d6): δ 163.0, 161.8, 161.2, 160.4, 160.1, 151.2, 147.9, 147.4, 144.1, 136.0, 132.9, 129.1, 128.9, 127.5, 126.4, 124.9, 118.4, 114.8, 109.6, 55.8; HRMS (+): calcd for C24H21N6O2 [M + H]+ 425.1721, found 425.1717, calcd for C24H20N6O2Na [M + Na]+ 447.154, found 447.1539.
10i: Yield: 82%; Rf, 0.45(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.79 (brs, 1H), 10.16 (s, 1H), 8.80–8.75 (m, 2H), 8.73 (d, J = 4.9 Hz, 1H), 8.46 (d, J = 7.9 Hz, 1H), 8.08 (dt, J = 1.6, 7.70 Hz, 1H), 8.04–8.00 (m, 2H), 8.00–7.95 (m, 2H), 7.82 (d, J = 4.9 Hz, 1H), 7.60 (ddd, J = 1.0, 4.8, 7.4 Hz, 1H), 7.50 (d, J = 7.0 Hz, 1H), 7.17–7.10 (m, 2H), 3.85 (s, 3H), 3.82 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.4, 163.1, 160.2, 160.2, 153.9, 153.1, 150.2, 148.4, 144.3, 143.0, 138.1, 129.0, 128.4, 126.3, 126.0, 124.8, 121.6, 118.7, 118.3, 117.5, 114.5, 109.3, 61.7, 56.2; HRMS (+): calcd for C25H23N6O3 [M + H]+ 455.1826, found 455.1828, calcd forC25H22N6O3Na [M + Na]+ 477.1646, found 477.1649.
10j: Yield: 84%; Rf, 0.42(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.63 (s, 1H), 10.15 (s, 1H), 8.77–8.79 (m, 1H), 8.72 (d, J = 4.9 Hz, 1H), 8.46 (d, J = 7.9 Hz, 1H), 8.40 (s, 1H), 8.08 (dt, J = 1.6, 7.7 Hz, 1H),8.03–7.99 (m, 2H), 7.95 (d, J = 8.8 Hz, 2H), 7.82 (d, J = 5.1 Hz, 1H), 7.60 (ddd, J = 1.0, 4.7, 7.5 Hz, 1H), 7.36 (s, 1H), 7.21 (d, J = 7.7 Hz, 1H), 7.04 (d, J = 8.2 Hz, 1H), 3.84 (s, 3H), 3.82 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.4, 163.1, 160.2, 160.2, 153.9, 151.1, 150.2, 149.5, 147.8, 144.5, 138.1, 129.0, 127.7, 126.3, 122.2, 121.6, 118.3, 111.9, 109.2, 108.6, 56.0, 55.9; HRMS (+): calcd for C25H23N6O3 [M + H]+ 455.1826, found 455.1830, calcd for C25H22N6O3Na [M + Na]+ 477.1646, found 477.1650.
10k: Yield: 88%; Rf, 0.65(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.84 (brs, 1H), 10.17 (s, 1H), 9.46 (d, J = 8.6 Hz, 1H), 9.19 (s, 1H), 8.78 (d, J = 4.3 Hz, 1H), 8.74 (d, J = 4.9 Hz, 1H), 8.47 (d, J = 7.9 Hz, 1H), 8.11–8.01 (m, 6H), 7.92 (d, J = 7.9 Hz, 1H), 7.82 (d, J = 4.9 Hz, 1H), 7.64–7.58 (m, 2H), 7.54 (d, J = 9.2 Hz, 1H), 7.45 (t, J = 7.1 Hz, 1H), 4.05–4.01 (m, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.4, 163.0, 160.3, 160.2, 158.2, 154.0, 150.2, 145.1, 144.3, 138.1, 133.0, 131.2, 129.3, 129.1, 128.9, 128.4, 126.5, 126.3, 126.2, 124.5, 121.1, 118.3, 115.0, 113.9, 109.2, 57.2; HRMS (+): calcd for C28H23N6O2 [M + H]+ 475.1877, found 475.1881, calcd for C28H22N6O2Na [M + Na]+ 497.1696, found 497.1700.
10l: Yield: 82%; Rf, 0.75(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.69 (brs, 1H), 10.16 (s, 1H), 8.80–8.76(m, 1H), 8.72 (d, J = 4.9 Hz, 1H), 8.45 (d, J = 7.9 Hz, 1H), 8.36 (brs, 1H), 8.08 (dt, J = 1.7, 7.7 Hz, 1H), 8.01 (d, J = 8.6 Hz, 2H), 7.93 (d, J = 8.8 Hz, 2H), 7.86 (s, 1H), 7.82 (d, J = 4.9 Hz, 1H), 7.60 (ddd, J = 1.1, 4.8, 7.5 Hz, 1H), 6.93 (d, J = 2.6 Hz, 1H), 6.65 (dd, J = 1.6, 3.0 Hz, 1H);13C NMR (150 MHz, DMSO‑d6): δ 163.4, 163.1, 160.2, 160.2, 153.9, 150.2, 150.1, 145.5, 144.3, 138.2, 137.3, 129.0, 126.3, 126.0, 121.6, 118.3, 113.6, 112.6, 109.3; HRMS (+): calcd for C21H17N6O2 [M + H]+ 385.1408, found 385.1408, calcd for C21H16N6O2Na [M + Na]+ 407.1227, found 407.1227.
10m: Yield: 84%; Rf, 0.78(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.70 (brs, 1H), 10.16 (s, 1H), 8.78 (dd, J = 0.7, 4.6 Hz, 1H), 8.72 (d, J = 4.9 Hz, 1H), 8.69 (brs, 1H), 8.45 (d, J = 7.9 Hz, 1H), 8.08 (dt, J = 1.6, 7.7 Hz, 1H), 8.01 (d, J = 8.6 Hz, 2H), 7.93 (d, J = 8.8 Hz, 2H), 7.82 (d, J = 4.9 Hz, 1H), 7.67 (d, J = 4.8 Hz, 1H), 7.60 (ddd, J = 1.1, 4.8, 7.5 Hz, 1H), 7.47 (d, J = 2.9 Hz, 1H), 7.15 (dd, J = 3.9, 4.8 Hz, 1H); 13C NMR (150 MHz, DMSO‑d6): δ 163.4, 163.0, 160.2, 160.2, 153.9, 150.2, 144.3, 142.7, 139.8, 138.1, 131.1, 129.2, 129.0, 128.3, 126.3, 126.1, 121.6, 118.3, 109.3; HRMS (+): calcd for C21H17N6OS [M + H]+ 401.1179, found 401.1179, calcd for C21H16N6OSNa [M + Na]+ 423.0999, found 423.0998.
10n: Yield: 85%; Rf, 0.79(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.93 (brs, 1H), 10.18 (s, 1H), 8.88 (brs, 1H), 8.78 (d, J = 4.0 Hz, 1H), 8.73 (d, J = 4.9 Hz, 1H), 8.63 (d, J = 4.0 Hz, 1H), 8.53 (brs, 1H), 8.46 (d, J = 7.9 Hz, 1H), 8.17 (d, J = 5.9 Hz, 1H), 8.08 (dt, J = 1.6, 7.7 Hz, 1H), 8.05–8.01 (m, 2H), 7.97 (d, J = 8.6 Hz, 2H), 7.82 (d, J = 4.9 Hz, 1H), 7.62–7.59 (m, 1H), 7.52 (dd, J = 5.0, 7.2 Hz, 1H); 13C NMR (150 MHz, DMSO‑d6): δ 163.4, 163.3, 160.2, 160.2, 153.9, 150.9, 150.2, 149.1, 144.7, 144.5, 138.1, 133.9, 130.9, 129.1, 126.3, 125.6, 124.5, 121.6, 118.3, 109.3; HRMS (+): calcd for C22H18N7O [M + H]+ 396.1567, found 396.1569, calcd for C22H17N7ONa [M + Na]+ 418.1387, found 418.1386.
11a: Yield: 85%; Rf, 0.35(CH3OH/CH2Cl2 = 1:20); 1H NMR(600 MHz, DMSO‑d6): δ 11.32 (brs, 1H), 10.22 (s, 1H), 8.91 (d, J = 6.2 Hz, 2H), 8.78 (d, J = 5.1 Hz, 1H), 8.31 (d, J = 6.2 Hz, 2H), 7.95 (d, J = 8.6 Hz, 2H), 7.88 (d, J = 8.6 Hz, 2H), 7.76 (brs, 1H), 7.69 (d, J = 5.1 Hz, 1H), 2.37–2.21 (m, 2H), 1.07 (t, J = 7.4 Hz, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 161.2, 160.7, 160.5, 160.4, 149.3, 149.1, 145.0, 129.2, 128.9, 122.2, 122.1, 118.5, 118.4, 110.4, 110.1, 25.9, 11.2; HRMS (+): calcd for C19H19N6O [M + H]+ 347.1615 found 347.1614, calcd for C19H18N6ONa [M + Na]+ 369.1434, found369.1433.
11b: Yield: 80%; Rf, 0.40(CH3OH/CH2Cl2 = 1:20); 1H NMR (600 MHz, DMSO‑d6): δ 11.32 (brs, 1H), 10.22 (s, 1H), 8.86–8.94 (m, 2H), 8.78 (d, J = 5.1 Hz, 1H), 8.30 (d, J = 6.2 Hz, 2H), 7.95 (d, J = 8.6 Hz, 2H), 7.88 (d, J = 8.6 Hz, 2H), 7.77–7.72 (m, 1H), 7.69 (d, J = 4.9 Hz,1H), 2.25 (q, J = 6.8 Hz, 2H), 1.59–1.47 (m, 2H), 0.94 (t, J = 7.3 Hz, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 161.1, 160.7, 160.5, 160.4, 151.9, 149.1,148.9, 146.6, 143.8, 129.2, 128.9, 122.3, 122.3, 118.5, 118.4, 110.1, 34.4, 20.0,14.1; HRMS (+): calcd for C20H21N6O [M + H]+ 361.1771, found 361.1770, calcd for C20H20N6ONa [M + Na]+ 383.1591, found 383.1589.
11c: Yield: 85%; Rf, 0.39(CH3OH/CH2Cl2 = 1:20); 1H NMR (600 MHz, DMSO‑d6): δ 11.76 (brs, 1H), 10.27 (s, 1H), 8.95–8.89(m, 2H), 8.79 (d, J = 5.1 Hz, 1H), 8.48 (brs, 1H), 8.31 (d, J = 6.2 Hz, 2H), 8.02–7.98 (m, 2H), 7.98–7.94 (m, 2H), 7.74 (d, J = 6.6 Hz, 2H), 7.70 (d, J = 5.1 Hz, 1H), 7.51–7.42 (m, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.1, 160.9, 160.8, 160.5, 159.0, 158.8, 148.5, 147.6, 147.1, 144.0, 135.0, 130.4, 129.3, 129.0, 127.5, 126.4, 122.5, 118.5, 110.3; HRMS (+): calcd for C23H19N6O [M + H]+ 395.1615, found 395.1614, calcd for C23H18N6ONa [M + Na]+ 417.1434, found 417.1433.
11d: Yield: 90%; Rf, 0.34(CH3OH/CH2Cl2 = 1:20); 1H NMR (600 MHz, DMSO‑d6): δ 11.70 (brs, 1H), 10.26 (s, 1H), 8.91 (d, J = 6.0 Hz, 2H), 8.79 (d, J = 5.0 Hz, 1H), 8.43 (s, 1H), 8.31 (d, J = 6.0 Hz, 2H), 8.01–7.97 (m, 2H), 7.96–7.93 (m, 2H), 7.70 (d, J = 4.9 Hz, 1H), 7.63 (d, J = 7.5 Hz, 2H), 7.28 (d, J = 7.9 Hz, 2H), 2.35 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.1, 160.9, 160.8, 160.5, 159.0, 158.8, 148.4, 147.7, 147.2, 143.9, 140.2, 132.2, 129.9, 129.0, 127.5, 126.5, 122.6, 118.5, 110.3, 21.5; HRMS (+): calcd for C24H21N6O [M + H]+ 409.1771, found 409.1771, calcd for C24H20N6ONa [M + Na]+ 431.1591,found 431.1590.
11e: Yield: 88%; Rf, 0.55(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.90 (brs, 1H), 10.26 (s, 1H), 8.88 (d, J = 6.0 Hz, 2H), 8.78 (d, J = 5.0 Hz, 1H), 8.72 (brs, 1H), 8.21–8.28 (m,2H), 8.04–8.00 (m, 2H), 7.99–7.94 (m, 3H), 7.68 (d, J = 4.9 Hz, 1H), 7.49 (q, J = 6.3 Hz, 1H), 7.34–7.28 (m, 2H); 13C NMR (150 MHz, DMSO‑d6): δ 163.1, 161.3, 160.7, 160.4, 161.2 (d,J = 249.8 Hz),149.4, 146.0, 144.2, 140.2,132.2(d,J = 8.8 Hz), 129.1, 126.7, 126.1, 125.4 (d,J = 3.8 Hz), 125.4, 122.5, 122.1,118.5, 116.5 (d, J = 20.9 Hz), 110.2; HRMS (+): calcd for C23H18FN6O [M + H]+ 413.1521, found 413.1521, calcd for C23H17FN6ONa [M + Na]+ 435.134, found 435.1342.
11f: Yield: 89%; Rf, 0.37(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ11.56 (brs, 1H), 10.24 (s, 1H), 8.92 (d, J = 5.0 Hz, 2H), 8.78 (d, J = 5.0 Hz, 1H), 8.36 (s, 1H), 8.32 (d, J = 6.0 Hz, 2H), 8.01–7.96 (m, 2H), 7.95–7.91 (m, 2H), 7.69 (d, J = 5.1 Hz, 1H), 7.57 (d, J = 8.3 Hz, 2H), 6.85 (d, J = 8.4 Hz, 2H); 13C NMR (150 MHz, DMSO‑d6): δ162.9, 160.9, 160.8, 160.5, 159.8, 159.0, 158.7, 148.5, 148.0,147.1, 143.8, 129.2, 128.9, 126.7, 125.9, 122.6, 118.5, 116.2, 110.2; HRMS (+): calcd for C23H19N6O2 [M + H]+ 411.1564 found 411.1562, calcd for C23H18N6O2Na [M + Na]+ 433.1383 found 433.1382.
11g: Yield: 86%; Rf, 0.13(CH3OH/CH2Cl2 = 1:5); 1H NMR (600 MHz, DMSO‑d6): δ 11.91 (brs, 1H), 10.24 (s, 1H), 8.84 (d, J = 4.6 Hz, 2H), 8.76 (d, J = 4.2 Hz, 1H), 8.52 (brs, 1H), 8.18 (d, J = 5.0 Hz, 2H), 8.05–7.99 (m, 4H), 7.96 (d, J = 7.9 Hz, 2H), 7.86 (d, J = 7.0 Hz, 2H), 7.66 (d, J = 4.4 Hz, 1H); 13C NMR (150 MHz, DMSO‑d6): δ 167.4, 162.2, 160.3, 160.3, 160.0, 152.1, 148.7, 146.3, 139.0, 135.0, 132.5, 132.0, 130.3, 129.1, 128.3, 127.5, 125.9, 124.5, 118.4, 109.6; HRMS (−): calcd for C24H17N6O3 [M−H]- 437.1938, found 437.1940.
11h: Yield: 88%; Rf, 0.57(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.63 (brs, 1H), 10.24 (s, 1H), 8.95–8.86 (m, 2H), 8.78 (d, J = 5.1 Hz, 1H), 8.41 (s, 1H), 8.27 (d, J = 6.2 Hz, 2H), 8.01–7.97 (m, 2H), 7.96–7.92 (m, 2H), 7.69 (d, J = 5.0 Hz, 2H), 7.68 (brs, 1H), 7.03 (d, J = 8.6 Hz, 2H), 3.82 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.0, 161.2, 161.1, 160.7, 160.5, 149.1, 147.5, 146.4, 143.9, 129.1, 129.0, 127.5, 126.6, 122.3, 118.5, 114.8, 110.2, 55.7; HRMS (+): calcd for C24H21N6O2 [M + H]+ 425.1721, found 425.1721, calcd for C24H20N6O2Na [M + Na]+ 447.154, found 447.1541.
11i: Yield: 84%; Rf, 0.47(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.80 (brs, 1H), 10.25 (s, 1H), 8.90 (d, J = 6.1 Hz, 2H), 8.78 (d, J = 5.1 Hz, 1H), 8.76 (s, 1H), 8.32–8.18 (m, 2H), 8.03–7.93 (m, 4H), 7.69 (d, J = 5.0 Hz, 1H), 7.50 (d, J = 7.0 Hz, 1H), 7.18–7.06 (m, 2H), 3.85 (s, 3H), 3.81 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.0, 160.9, 160.8, 160.5, 159.1, 158.7, 153.1, 148.4, 148.0,144.0, 143.1, 129.0, 128.4, 126.4, 124.8, 122.7, 118.5, 117.5,114.5, 110.3, 617, 56.2; HRMS (+): calcd for C25H23N6O3 [M + H]+ 455.1826, found 455.1827, calcd for C25H22N6O3Na [M + Na]+ 477.1646, found 477.1647.
11j: Yield: 87%; Rf, 0.49(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz,DMSO‑d6): δ 11.62 (s,1H), 10.20 (s, 1H), 8.79–8.86 (m, 2H), 8.74 (d, J = 5.1 Hz, 1H), 8.40 (s, 1H), 8.17–8.11(m, 2H),8.04–7.97(m, 2H), 7.96–7.91 (m, 2H), 7.64 (d, J = 5.1 Hz, 1H), 7.35 (s, 1H),7.21 (d, J = 8.1 Hz, 1H), 7.04 (d, J = 8.3 Hz, 1H), 3.84 (s, 3H), 3.82 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.0, 162.0, 160.5, 160.5, 151.1,149.5,147.8, 144.2, 144.0, 128.9, 127.7, 126.5, 122.2, 121.4, 118.4, 112.0, 109.8, 108.7, 56.0, 55.9; HRMS (+): calcd for C25H23N6O3 [M + H]+ 455.1826, found 455.1828, calcd for C25H22N6O3Na [M + Na]+ 477.1646, found 477.1649.
11k: Yield: 88%; Rf, 0.77(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.85 (s, 1H), 10.22 (s, 1H), 9.44 (d, J = 8.6 Hz, 1H), 9.18 (s, 1H), 8.83 (d, J = 5.0 Hz, 2H), 8.75 (d, J = 5.1 Hz, 1H), 8.15 (d, J = 6.0 Hz, 2H), 8.06 (d, J = 9.2 Hz, 1H), 8.02(s, 4H), 7.92 (d, J = 7.9 Hz, 1H), 7.64 (d, J = 5.1 Hz, 1H), 7.60 (t, J = 7.5 Hz, 1H), 7.54 (d, J = 9.2 Hz, 1H), 7.45 (t, J = 7.15 Hz, 1H), 4.03 (s, 3H); 13C NMR (150 MHz, DMSO‑d6): δ 163.1, 161.9, 160.5, 160.5, 158.2, 150.9, 145.2, 133.0, 131.2, 129.3, 129.1, 128.9, 128.5, 126.5, 126.3, 124.5, 121.5, 118.4, 114.9, 113.9, 109.9, 57.2; HRMS (+): calcd for C28H23N6O2 [M + H]+ 475.1877, found 475.1882, calcd for C28H22N6O2Na [M + Na]+ 497.1696, found 497.1700.
11l: Yield: 88%; Rf, 0.82(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.71 (brs, 1H), 10.27 (s, 1H), 8.92 (d, J = 6.2 Hz, 2H), 8.79 (d, J = 5.0 Hz, 1H), 8.37 (brs, 1H), 8.32 (d, J = 6.2 Hz, 2H), 8.02–7.97(m, 2H), 7.95–7.92 (m, 2H), 7.86 (s, 1H), 7.71 (d, J = 4.9 Hz, 1H), 6.93 (d, J = 2.4 Hz, 1H), 6.65 (d, J = 1.3 Hz, 1H); 13C NMR(150 MHz, DMSO‑d6): δ 163.1, 160.8, 160.4, 159.0, 158.8, 150.1, 148.4, 147.2, 145.5, 144.0, 137.4, 129.0, 126.3, 122.6, 118.5, 113.6, 112.6, 110.3; HRMS (+): calcd for C21H17N6O2 [M + H]+ 385.1408, found 385.1406, calcd for C21H16N6O2Na [M + Na]+ 407.1227, found 407.1226.
11m: Yield: 86%; Rf, 0.79(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 11.72 (brs, 1H), 10.27 (s, 1H), 8.92 (d, J = 6.2 Hz, 2H), 8.79 (d, J = 5.1 Hz, 1H), 8.69 (s, 1H), 8.32 (d, J = 6.2 Hz, 2H), 8.02–7.97 (m, 2H), 7.96–7.91(m, 2H), 7.70 (d, J = 5.0 Hz, 1H), 7.67 (d, J = 4.6 Hz, 1H), 7.47 (d, J = 2.9 Hz, 1H), 7.18–7.13 (m, 1H); 13C NMR (150 MHz, DMSO‑d6): δ 163.0, 160.8, 160.5, 159.0, 158.8, 148.4, 147.2, 144.0, 142.8, 139.8, 131.1, 129.2, 129.0, 128.3, 126.4, 122.6, 118.5, 110.3; HRMS (+): calcd for C21H17N6OS [M + H]+ 401.1179, found 401.1180, calcd for C21H16N6OSNa [M + Na]+ 423.0999, found 423.0998.
11n: Yield: 87%; Rf, 0.82(CH3OH/CH2Cl2 = 1:10); 1H NMR (600 MHz, DMSO‑d6): δ 12.02 (brs, 1H), 10.29 (s, 1H), 8.88–8.95 (m, 2H), 8.79 (d, J = 5.1 Hz, 1H), 8.65 (d, J = 4.8 Hz, 1H), 8.51 (brs,1H), 8.35–8.26 (m, 2H), 8.06–8.01 (m, 3H), 8.00–7.93 (m, 3H), 7.70 (d, J = 5.0 Hz, 1H), 7.50–7.45 (m, 1H); 13C NMR (150 MHz, DMSO‑d6): δ 161.1, 160.8, 160.4, 159.0, 158.7, 153.3, 149.3, 148.9, 146.9, 146.7, 144.3, 138.2, 129.2, 126.0, 125.0, 122.4, 120.8, 118.5, 117.3, 110.3; HRMS (+): calcd for C22H18N7O [M + H]+ 396.1567, found 396.1568, calcd for C22H17N7ONa [M + Na]+ 418.1387, found 418.1387.
4.2 Biological evaluation
4.2.1 Luciferase assay
Luciferase detection was performed in NH2 cells as described previously. NH2 cells were seeded in 6-well plates and treated with targets compounds or flavanol for 8 h. Then test the luciferase activity by luciferase kit of Promega E1500.
4.2.2 MTT assay
All cancer cells used for the MTT assay were cultured in DMEM medium (ThermoFisher Scientific) containing 10% fetal bovine serum. The cells were cultured in 96-well plates with 100 µL DMEM, and the total amount of each well was about 1 × 104 when the compound was added. Each compound was gradient treated at six concentrations with six replicates per treatment. Meanwhile, dimethyl sulfoxide (DMSO) was used as negative control. After 48 h, 10 µL MTT solution (5 mg/mL, M2128, Sigma) was added to each well and cultured in a cell incubator for another 4 h. Discard the medium of each well, add 100 µL DMSO, and dissolve. After catching sharks for a short time, the light intensity was detected at 490 nm, and the IC50 value was calculated using GraphPad prism5 software (GraphPad Software, San Diego, CA, USA).
4.2.3 Western blot analysis
Cells were harvested after compounds treatment, and lysed with 1 × SDS loading buffer. Then the samples were boiled for western blot analysis with indicated primary antibodies, RNA Pol II CTD P-Ser2 (A300-654A, Bethyl), Tubulin (T5168, Sigma), PARP1 (9542, CST, Shanghai, China).
4.2.4 Annexin-V-FITC apoptosis assay
Cells were cultured in a 6-well plate (about 1 × 105 in each well) and treated with different concentrations of 9 h for 24 h. Cells were then collected and analyzed for apoptosis as planned (BD Bioscience, Franklin Lakes, NJ, USA). The apoptosis status was analyzed by flow cytometry, and the data were analyzed by FlowJo7.6 software.
4.3 Molecular docking
The crystal structure of the CDK9/cyclinT1 complex (3BLR) was retrieved from the PDB database (https://www.rcsb.org/structure/3BLR) with a resolution of 2.80 Å. Schrödinger software (Version 2020-3) was used to perform the docking study. Briefly, the structure of 9 h and the native ligand (FLP) of CDK9 were prepared by the LigPrep (Version 5.5) of Schrödinger with default settings. The Protein Preparation Wizard module dealt with the crystal structure of CDK9. In this process, waters were removed, hydrogen atoms were added, missing side chains and loops were filled using Prime (Version 6.1), hydrogen bonds were assigned, and the complex was subjected to constrained minimization by the force field of OPLS3e (Roos et al., 2019). The Induced Fit Docking panel was used to investigate the possible binding model of 9 h. Firstly, FLP was redocked into the active pocket of CDK9 to validate the accuracy of the docking method by the standard induced-fit docking protocol (Sherman et al., 2006a, 2006b) with default parameters. Subsequently, the result gave a heavy atom ligand root‐mean‐square deviation (RMSD) of 0.1755 Å between the native ligand and the redocked FLP (Fig. S4). In addition, the binding free energy of the docking models were calculated by MM/GBSA (Prime MMGBSA v3.000) in an implicit solvent. Schrödinger's Maestro (Version 12.5) was applied as the primary graphical user interfaces for the visualization of the crystal structure and docking results. The figures of the molecular docking results were analyzed and generated by PyMol (The PyMOL Molecular Graphics System, Version 2.3 Schrödinger, LLC.) (Fig. S4).
Acknowledgements
This work was supported by grants from the Fundamental Research Funds for the Central Universities (No. 20720180051), Zhejiang basic public welfare fund (No. LGF22H300015), and Natural Science Foundation of Ningbo (Grant No. 202003N4160).
Author contributions
Hongyu Hu, Fengming He, and Shengxian Zhao synthesized all target compounds and confirmed their structure. Cong Wang, Cao Yin, and Chenfan Li carried out biological experiments. Hongyu Hu, Fengming He, and Cong Wang prepared the original manuscript. Zhen Wu and Meijuan Fang designed the research and revised the paper.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- The cyclin-dependent protein kinases and the control of cell division. FASEB J.. 1994;8:1114-1121.
- [CrossRef] [Google Scholar]
- Synthesis, structure–activity relationship studies and biological evaluation of novel 2,5-disubstituted indole derivatives as anticancer agents. Chem. Biol. Drug Des.. 2016;88:766-778.
- [CrossRef] [Google Scholar]
- Design, synthesis and biological evaluation of methylenehydrazine-1-carboxamide derivatives with (5-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)-1H-indole scaffold: Novel potential CDK9 inhibitors. Bioorg. Chem.. 2020;102:104064
- [CrossRef] [Google Scholar]
- CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev.. 2014;28:1800-1814.
- [CrossRef] [Google Scholar]
- Flavopiridol in Untreated or Relapsed Mantle-Cell Lymphoma: Results of a Phase II Study of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol.. 2003;21:1740-1745.
- [CrossRef] [Google Scholar]
- The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res.. 2013;41:277-287.
- [CrossRef] [Google Scholar]
- Viral-host interactions that control HIV-1 transcriptional elongation. Chem. Rev.. 2013;113:8567-8582.
- [CrossRef] [Google Scholar]
- Identification of Atuveciclib (BAY 1143572), the First Highly Selective, Clinical PTEFb/CDK9 Inhibitor for the Treatment of Cancer. ChemMedChem. 2017;12:1776-1793.
- [CrossRef] [Google Scholar]
- Synthesis, anti-tuberculosis activity, and 3D-QSAR study of 4-(adamantan-1-yl)-2-substituted quinolines. Bioorg. Med. Chem.. 2007;15:626-640.
- [CrossRef] [Google Scholar]
- Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat. Chem. Biol.. 2018;14:163-170.
- [CrossRef] [Google Scholar]
- Design, synthesis and biological activities of Nilotinib derivates as antitumor agents. Bioorg. Med.Chem.. 2013;21:2527-2534.
- [CrossRef] [Google Scholar]
- Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem.. 1998;273:13855-13860.
- [CrossRef] [Google Scholar]
- Targeting CDK9 for treatment of colorectal cancer. Mol. Oncol.. 2019;13:2178-2193.
- [CrossRef] [Google Scholar]
- OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput.. 2019;15:1863-1874.
- [CrossRef] [Google Scholar]
- Flavopiridol: the First Cyclin-Dependent Kinase Inhibitor in Human Clinical Trials. Invest. New Drugs. 1999;17:313-320.
- [CrossRef] [Google Scholar]
- Use of an Induced Fit Receptor Structure in Virtual Screening. Chem. Biol. Drug Des.. 2006;67:83-84.
- [CrossRef] [Google Scholar]
- Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J. Med. Chem.. 2006;49:534-553.
- [CrossRef] [Google Scholar]
- A novel Cdk9 inhibitor preferentially targets tumor cells and synergizes with fludarabine. Oncotarget. 2014;5:375-385.
- [CrossRef] [Google Scholar]
- Recent Developments in the Biology and Medicinal Chemistry of CDK9 Inhibitors: An Update. J. Med. Chem.. 2020;63:13228-13257.
- [CrossRef] [Google Scholar]
- Antitumor action of CDK inhibitor LS-007 as a single agent and in combination with ABT-199 against human acute leukemia cells. Acta Pharmacol. Sin.. 2016;37:1481-1489.
- [CrossRef] [Google Scholar]
- A Novel CDK9 Inhibitor Shows Potent Antitumor Efficacy in Preclinical Hematologic Tumor Models. Mol. Cancer Ther.. 2014;13:1442.
- [CrossRef] [Google Scholar]
- RNA polymerase II elongation control. Annu. Rev. Biochem.. 2012;81:119-143.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.104039.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







