Development and validation of a two-step assay for differentiation of Penis et testis cervi from cervi nippon Temmink and cervi elaphus Linnaeus based on specific-species polymerase chain reaction-restriction fragment length polymorphism patterns
⁎Corresponding authors at: School of Laboratory Medicine, Beihua University, No.3999 East Road of Binjiang, Jilin, Jilin 132013, China. aijinxia@beihua.edu.cn (Jinxia Ai), limingcheng@beihua.edu.cn (Mingcheng Li)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University. Production and hosting by Elsevier.
Abstract
Penis et testis cervi is one of the precious traditional Chinese medicines (TCMs) in China, which, when authentic, is sourced from species of Cervi nippon Temmink (C. Nippon) and Cervi elaphus Linnaeus (C. elaphus L). Authentication of species of Penis et testis cervi remains technically challenging. This study attempted to modify the species-specific polymerase chain reaction (PCR) and PCR-restriction fragment length polymorphism (PCR-RFLP) method using restriction enzyme Hpy188III for the identification of Penis et testis cervi species. First, the cytochrome b (Cyt b) and cytochrome c oxidase subunit I (COI) genes in mitochondrial DNA from two authentic species and their counterfeits or adulterants were downloaded and analyzed with bioinformatics tools to establish the phylogenetic tree, then a series of primers were designed, and the restriction enzyme sites were confirmed. Genomic DNA was extracted from all samples by the improved SDS method. Species-specific PCR reactions were optimized to differentiate authentic Sika deer and Red deer Penis et testis cervi from their counterfeits or adulterants. PCR-RFLP using single restriction enzyme Hpy188III was performed in Cyt b regions to identify the species of Sika deer and Red deer. Simultaneously, recombination plasmid carrying the cyt b fragment could be cloned, sequenced, and blasted. The assay specificity and sensitivity were evaluated, and 30 batches of commercial samples were validated. The phylogenetic tree showed that the physico-chemical properties of Sika deer and Red deer variants were similar. Sufficient DNA extracts were extracted from all samples used in the improved SDS method. Establishment and optimization of a species-specific PCR reactions and PCR-RFLP system were undertaken to identify deer species in Penis et testis cervi samples. A specific PCR-RFLP pattern with three distinct products of sizes 155, 113, and 204 bp was exhibited in Sika deer species, and two distinct products of 204 and 268 bp in size were evident in Red deer species, which were significantly different from the pattern of the counterfeits or adulterants. The distinctive PCR-RFLP fingerprint pattern using the restriction enzyme Hpy188III is a rapid and reliable assay for identification of the Penis et testis cervi species.
Keywords
Authenticity
Cervi nippon Temmink
Cervi elaphus Linnaeus
PCR-RFLP
Penis et testis cervi
Sika deer and Red deer
1 Introduction
Penis et testis cervi, also known as “Lu Shen” is one of the precious traditional Chinese medicines (TCMs) used in China. It is made up of two parts, dried deer testis and penis including glands, sperm cells in addition to muscle and skin (Tang et al.,2002). There are abundant deer species: the testis and penis from Sika deer (Cervi nippon Temmink, C. Nippon) and Red deer (Cervi elaphus Linnaeus, C. elaphus L) were recorded as authentic medicinal materials according to China Pharmacopoeia (2010 edition) (Chinese Pharmacopoeia Commission, 2010). Other species were recorded as adulterants (different subspecies of deer) or counterfeits (different animal species). Although both are authentic medicinal materials, their medicinal functions are different. Some studies have shown that the water extract of Sika deer testis and penis is better than the water extract of Red deer in protecting mice from cisplatin-induced acute renal injury (Wang et al.,2016), while the anti-fatigue effect of Red deer testis and penis is higher than that of Sika deer (Wang et al.,2017).
In addition, the prices of Sika deer and Red deer testis and penis in the Chinese medicine market also differ greatly, with Sika deer testis and penis being the most precious, commanding the highest price. Due to the different medicinal values and price difference of the two kinds of testis and penis, driven by economic interests, the phenomenon of mislabeling Red deer testis and penis as Sika deer (when offered for sale) often occurs in the market, which not only infringes the legitimate rights and interests of consumers, but also affects the normal operation of the TCM market (Tang et al.,2002). Therefore, identification of their authenticity, animal species, or origins become key to controlling such herbal products. There is an urgent need for an efficient and accurate method to identify Sika deer and Red deer testis and penis, ensuring its clinical efficacy, and providing a scientific basis for the identification and analysis of Sika deer and Red deer testis and penis. Traditional identification methods of TCM materials include identification of origin, identification of characteristics, microscopic identification, and physico-chemical identification, but these identification techniques are easily affected by high-temperature processing, subjective factors, and other extraneous influences (Zhang,2010; Noguchi et al., 2012; Cronin et al.,1991; Zhang et al., 2007). In recent years, with the development of molecular biological technology, polymerase chain reaction (PCR) has been widely used in the identification of TCM (Wang et al.,2019; Niu et al., 2021). Our team has established molecular biological identification methods to identify the authenticity of deer fetus, deer testis and penis based on the cytochrome b (Cyt b) gene in mitochondrial DNA (mt DNA), but these methods are mainly used for the identification of authenticity and cannot achieve species identification (Li et al., 2016; Ai et al., 2017); because the provenance of Sika deer testis and penis is similar to that of Red deer testis and penis, this study is based on PCR technology and Restriction Fragment Length Polymorphism (RFLP) can help identify the provenance of its species. The purpose is to establish a method for distinguishing Sika deer testis and penis and Red deer testis and penis based on PCR-RFLP technology, to supplement the current identification method of deer testis and penis, and to provide a reference for accurate detection and quality control thereof.
2 Materials and methods
2.1 Collection of samples
Four batches of authentic Penis et testis cervi were purchased from the Key Field Observation Experiment Station in the Changbai Mountains Natural Reserve of the Ministry of Agriculture and Rural Affairs. These authentic Penis et testis cervi included two from Sika deer species and two from Red deer species, which were used as standard reference samples originating from Sika deer (batch number: MHL-B211008-009) and from Red deer (batch number: ML-B211015-016). A total of five batches of 50 adulterant samples included Cervus Canadensis (New Zealand deer) and Rangifer tarandus (Reindeer); ten batches of 30 counterfeit samples consisted of pig, cattle, and sheep (Table 1). Commercially available deer Penis et testis cervi products included 30 batches of 60 samples labeled with Sika deer and Red deer species, respectively. They were vouched as being authentic Penis et testis cervi identified in morphology by Yinglan Jin (Director of the Food and Drug Supervision and Management, Jilin, China). All animal tissues used in the present study were used in compliance with the Animal Research: Reporting In Vivo Experiments (ARRIVE) Guidelines (Ethical approval number: Protocol Number 2021–09-10).
| Series No. | Sample’s species | Source | C(ng•μL−1) | Purity |
|---|---|---|---|---|
| MHL2110008 | Sika deer | Changbai Mountain, Jilin | 314.3 | 1.81 |
| MHL2110009 | Sika deer | Changbai Mountain, Jilin | 315.3 | 1.82 |
| ML21101015 | Red deer | Changbai Mountain, Jilin | 311.3 | 1.84 |
| ML21101016 | Red deer | Changbai Mountain, Jilin | 312.3 | 1.83 |
| XXL-01 | New Zealand deer | Changbai Mountain, Jilin | 248.9 | 1.91 |
| XL-01 | Reindeer | Changbai Mountain, Jilin | 213.7 | 1.87 |
| ZB-01 | Pig | Market, Jilin | 101.2 | 1.92 |
| NB-01 | Cattle | Market, Jilin | 103.6 | 1.81 |
| YB-01 | Sheep | Market, Jilin | 397.2 | 1.91 |
2.2 Analysis of deer testis and penis genome and designing a series of primers
Nucleotide sequences of Cyt b of five species of deer species including Red deer (Accession No. AB924664.1), Sika deer (Accession No. MN883859.1), white-tailed deer (Accession No. KX171728.1), reindeer (Accession No. DQ673135.1), roe deer (Accession No. Y14951.1), and four non-deer species: horse (Accession No. EU308145.1), goat (Accession No. AB044308.1), cattle (Accession No. MN148732.1), and dog (Accession No. KC985187.1) were downloaded from GenBank. Additionally, Sika deer cytochrome c oxidase subunit I (COI) gene (Accession No. HM204520.1) and Red deer COI gene (Accession No. HM204520.1) were included in the study. The phylogenetic tree was constructed based on the NJ (neighbor-joining) method, using Boot-step (set to repeat 1000 times) using Mega7.0 software and R software. Then ProtParam software in ExPASy was used to pRedict the relative molecular weight, amino acid contents, and other physico-chemical properties of the proteins encoded by Cyt b gene from Sika deer and Red deer. Meanwhile, DNAMAN software was used to analyze the restriction endonuclease map of the Cyt b gene of the Sika deer and the Red deer. The restriction endonuclease Hpy188III (5′… TC'NNGA… 3′) was screened according to the base difference between sequences, the specificity of restriction endonuclease recognition site, the size and number of digested fragments, and the degree of separation of the digested fragments on the agarose gel electrophoresis.
Sequence comparison was conducted using DNAMAN software to find the inter-specific and intra-specific differences. Finally, Primer 5.0 software was applied to design two pairs of primers.
Primer 1 (upstream: 5′-GCCAACCTGGTACTCTGCTTG-3′ and downstream: GATGATGC TAAAAG T AGTAAG-3′) was used to differentiate the authenticity of Sika deer and Red deer from counterfeits and adulterants.
Primer 2 (upstream:5′-TACCATGAGGACAAATATCATTCTG-3′and downstream: 5′-CCTCCTAGTTTGTTGGGAATTGATCG-3′) was applied in differentiation of species of Sika deer and Red deer. Both of primers were synthesized by Sangon Biotech Co., Ltd (Shanghai, China).
2.3 Extraction of genomic DNA from samples
All samples used in the study were washed with 75% ethanol, dried, cut into small pieces, put into a mortar, whereupon liquid nitrogen was added and the material ground to powder, and retained for later use. The improved SDS method (as optimized by our team) was used to extract the sample genomic DNA with reference to an animal genomic DNA extraction method (Duan et al., 2020). In a brief, we weighed about 0.05 g of the ground samples and placed it in an Ep tube. A P1 solution preheated at 37 ℃ (500 μL), P2 solution (30 μL), and P3 solution (15 μL) was added. We evenly mixed these, placed the samples in a water bath at 56 ℃ and shook the samples for 1 h. We added 500 μL P4 solution, centrifuged the samples at 10,000 rpm for 10 min, took 500 μL of the supernatant, added an equal volume of P5 solution, stoRed the samples at −20 ℃ for 1 h, and centrifuged it at 10,000 rpm for 10 min. Some 30 μL of P6 solution was removed by suction and the residue at the bottom of the pipe was rinsed twice, then centrifuged at 10,000 rpm for 10 min; the supernatant was discarded after centrifugation. The Ep tube was left open and placed on sterile filter paper, dried, and 80 μL sterilized double-distilled water was added: the sample DNA solution was obtained by dissolving DNA in sterilized double-distilled water, and stoRed at −20 ℃ for later analysis.
2.4 Identification of sample genomic DNA quality
The DNA concentration of the sample DNA solution obtained from the above operations was determined using an ultraviolet spectrophotometer (Q6000, Quawell, USA); each sample was analyzed three times, and the purity of the sample was calculated according to the A260/A280 ratio. Meanwhile, 5 μL of DNA solution and 6 × loading buffer solution 1 μL was evenly mixed, and the sample was subjected to electrophoresis on the 0.8% agarose gel electrophoresis plate containing nucleic acid dye (GelRed 0.5 mg/L). The voltage was 110 V, and the electrophoresis occurRed over 50 min. The sample was examined by a UV gel imaging analyzer and photographed.
2.5 Authentication of deer Penis et testis cervi with a species-specific PCR
PCR amplification was conducted with specific primers (Primer 1) for the authenticity of deer genera in Penis et testis cervi to exclude adulterants and counterfeits. The PCR reaction system amounted to 25 μL including 2 × Taq Master Mix 12.5 μL, 0.5 μL upstream primer, and 0.5 μL downstream primer respectively. The volume of DNA template was 0.8 μL and that of the double-distilled water was 10.7 μL. Reactions were performed in 96-well optical plates and run in a machine (T100 Thermal Cycler, Biorad, CA). Cycle parameters were as follows: pre-denatuRed at 94 ℃ for 5 min, 28 cycles (94 ℃, 30 s; 60 ℃, 30 s; 72 ℃, 30 s), extended at 72 ℃, 10 min, and stoRed at 4 ℃.
2.6 Establishment and optimization of PCR-RFLP system to identify deer species in Penis et testis cervi samples
The first step was PCR amplification of partial fragments from the authentic deer Penis et testis cervi, which was conducted with specific primers (Primer 2) and the PCR condition matched that used in the first step. The second step entailed the PCR-RFLP reaction. The enzyme digestion reaction system amounted to 20 μL, including restriction endonuclease (0.6 μL), 10 × NE Buffer (2 μL), plus PCR product (4 μL). This was replenished with sterilized double-distilled water to 20 μL, mixed and reacted at 37 ℃ for 15 min. The reaction products were mixed with 10 × loading buffer in the ratio of 5:1, and assayed using 2% agarose gel electrophoresis (GelRed® staining), and the gel imaging system recorded photographs for later analysis.
2.7 Cloning and verification of deer Penis et testis cervi sequences
To verify the accuracy of the identification assay, sequencing was conducted on some genuine deer Penis et testis cervi. The specific DNA fragment of deer Penis et testis cervi amplified by PCR was excised, purified with a QIAqick PCR purification kit, and ligated to pGEM-T easy vector (Promega, USA) then transformed into DH5α competent cells, and the white clones were screened. Nucleotide sequencing was performed directly on cloned fragments using an ABI Prism 377 DNA sequencer. Sequence similarity searching was conducted with the BLAST program available at the website of the National Center of Biotechnology Information (www.ncbi.nlm.nih.gov).
2.8 Evaluation of the assay on specificity and sensitivity
To validate the assay specificity, mimic counterfeit deer Penis et testis cervi containing Sika deer and Red deer species in the ratios of 1:90, 1:80, 1:70, 1:60, 1:50, 1:40, 1:30, 1:20, 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, and 1:3 (weight/weight) were premixed separately. Mixture DNA was extracted by the modified method as templates. Some 0.5 μL of the template was added to the 19.5-μL tubes according to the requirements of the PCR reaction system and PCR-RFLP amplification was performed as per the above method. To validate the assay detection sensitivity, the Sika deer Penis et testis cervi DNA extracts were diluted with double-distilled water to 100 ng/μL. The samples were unfolded along a 10-fold gradient to 0.001 ng based on the preliminary 0.1 ng of Sika deer Penis et testis cervi DNA extract, and 0.5 μL was taken as a template. PCR-RFLP amplification were conducted as per the aforementioned method.
2.9 Evaluation of commercially available samples
A total of 30 batches of 60 commercially available deer Penis et testis cervi products, which were labeled as deer Penis et testis cervi samples, the Red deer species samples were successively numbered as S1-15, the Sika deer as S16-30 were verified. The aforementioned optimized species-specific PCR and PCR-RFLP analysis system were performed to authenticate the identification of deer species among Penis et testis cervi samples.
3 Results
3.1 Bioinformatics analysis
The nucleotide sequences of Cyt b gene of Sika deer and Red deer were compaRed with those of seven different species registeRed in GenBank using Mega 7.0 software. The phylogenetic tree of Sika deer and Red deer could be divided into deer and non-deer families based on Cyt b nucleotide sequences, of which the Sika deer and Red deer were the closest, and the white-tailed deer and reindeer were clusteRed into one branch. The basic physico-chemical properties of the Cytb protein amino acid sequence of Sika deer were investigated using ProtParam and ProtScale software in ExPASy. The results showed that the physico-chemical properties of Sika deer and Red deer were also similar (Fig. 1, Table 2). Meanwhile, a specific restriction endonuclease recognition site for the Hpy188III (5′- TC^NNGA-3′) was only found at different positions in the Cytb region sequence of Sika deer and Red deer. The strategy in the selection of primer and restriction enzyme site between Sika deer and Red deer is illustrated in Fig. 2.
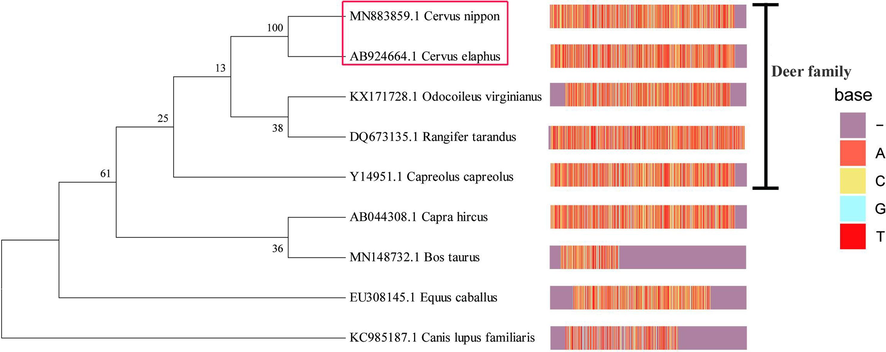
-
Phylogenetic tree based on Cyt b region derived from 5 species of deer family and 4 species of non-deer family. Footnote: This tree was constructed with Neighbor-Joining method. The left is the phylogenetic tree, and the right represents the nucleotide sequence of Cyt b gene of each species. Bootstrap after statistical analysis from 1,000 trees are shown on each node.
| Species | Molecular formula | Atomic number | Number of amino acid | Leucine contents | Isoleucine contents | Instability coefficient | Fat index |
|---|---|---|---|---|---|---|---|
| C. nippon | C2043H3094N470O502S19 | 6 218 | 379 | 14.8% | 10.8% | 42.06 | 120.9 |
| C. elaphus | C2040H3088N470O501S19 | 6 118 | 379 | 15.0% | 9.8% | 39.63 | 120.4 |
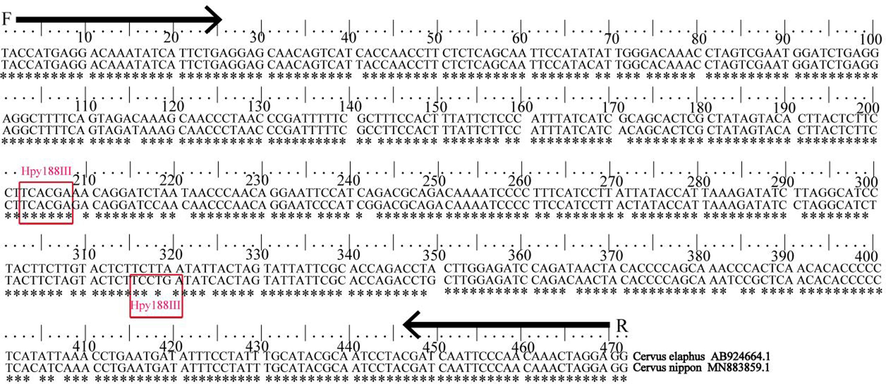
-
Strategy in the selection of primer and restriction enzyme site between Sika deer and Red deer. Footnote: Comparison of nucleotide sequence restriction sites of Cyt b gene primer region between Sika deer and Red deer. C. elaphus represents Red deer and C. nippon represents Sika deer; *represents the same base;F represents forward primer and R represents reverse primer;Red box represents the site for Hpy188III.
3.2 Analysis of DNA concentration, purity, and integrity
After repeated determination, the purity of the extracted genomic DNA from the deer Penis et testis cervi and their counterfeit samples was between 1.8 and 2.0, which proved that the DNA samples were free from protein contamination (Table 1). The genomic DNA extracted by the improved SDS method from deer Penis et testis cervi and other counterfeit samples can be found with clear, bright bands at about 23,130 bp (Fig. 3). This finding indicates that the extracted genomic DNA had undergone no degradation and was suitable for subsequent experiments.
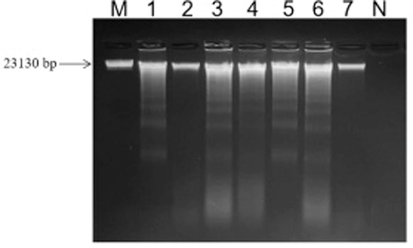
-
Agarose gel electrophoretogram of DNA extraction from deer Penis et testis cervi samples as well as adulterants and counterfeits. Footnote: M-λDNA Marker;1. MHL-B;2. ML-B;3. XXL-B;4. XL-B;5. ZB;6. NB;7. YB;N. Negative control. MHL-B, Chinese spelling name (Mei hua lu bian), represents authentic Sika deer Penis et testis cervi. ML-B, Chinese spelling name (Ma lu bian), represents authentic Red deer Penis et testis cervi. XXL-B, Chinese spelling name (Xin xi lan bian), represents New Zealand deer Penis et testis cervi. XL-B, Chinese spelling name (Xun lu bian), represents Reindeer Penis et testis cervi. ZB, Chinese spelling name (Zu bian), represents Pig Penis et testis cervi. NB, Chinese spelling name (Niu bian), represents Cattle Penis et testis cervi. YB, Chinese spelling name (Yang bian), represents Sheep Penis et testis cervi.
3.3 Optimization of PCR for authenticity of deer Penis et testis cervi
The authentic deer Penis et testis cervi, their adulterants and counterfeits were amplified by the optimized PCR reaction system for identification of authenticity of deer Penis et testis cervi. The results showed that the authentic deer Penis et testis cervi (both of Sika deer and Red deer) showed clear, bright bands at 223 bp, while the counterfeit and adulterated products showed no target bands, indicating that the designed primers were highly specific and could be used in subsequent experiments (Fig. 4a).
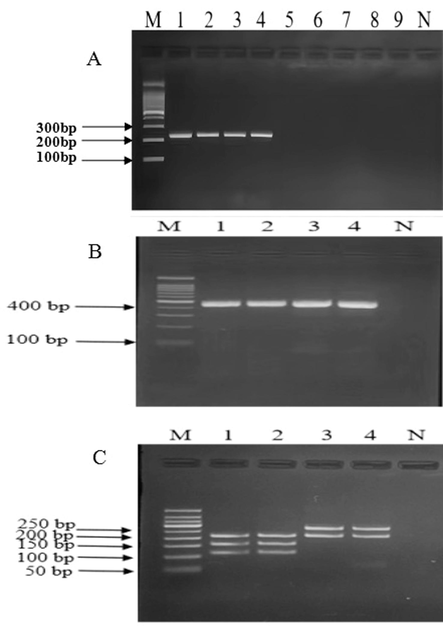
-
Agarose gel electrophoretogram of authenticity of deer Penis et testis cervi and identification of deer species by a species PCR and PCR-RFLP. A: Agarose gel electrophoretogram of authenticity of deer Penis et testis cervi by a species PCR. Footnote: M−100 bp Ladder Marker;1. MHL-B;2. MHL-B;3. ML-B;4. ML-B;5. XXL-B;6. XL-B;7. ZB;8. NB;9. YB;N. Negative control. MHL-B, Chinese spelling name (Mei hua lu bian), represents authentic Sika deer Penis et testis cervi. ML-B, Chinese spelling name (Ma lu bian), represents authentic Red deer Penis et testis cervi. XXL-B, Chinese spelling name (Xin xi lan bian), represents New Zealand deer Penis et testis cervi. XL-B, Chinese spelling name (Xun lu bian), represents Reindeer Penis et testis cervi. ZB, Chinese spelling name (Zu bian), represents Pig Penis et testis cervi. NB, Chinese spelling name (Niu bian), represents Cattle Penis et testis cervi. YB, Chinese spelling name (Yang bian), represents Sheep Penis et testis cervi. B: Agarose gel electrophoretogram of authentic deer Penis et testis cervi PCR products. Footnote: M−100 bp Ladder Marker;1. MHL-B;2. MHL-B;3. ML-B;4. ML-B;N. negative control. C: Agarose gel electrophoretogram of authentic deer Penis et testis cervi by PCR-RFLP. Footnote: M−100 bp Ladder Marker;1. MHL-B;2. MHL-B;3. ML-B;4. ML-B; N. Negative control. MHL-B, Chinese spelling name (Mei hua lu bian), represents authentic Sika deer Penis et testis cervi (1. MHL2110008; 2. MHL2110009). ML-B, Chinese spelling name (Ma lu bian), represents authentic Red deer Penis et testis cervi (3. ML21101015; 4. ML21101016).
3.4 Identification of deer species by PCR-RFLP
After RFLP analysis of PCR products, the Sika deer characteristic PCR-RFLP fingerprints exhibited three distinct fragments that appeared on the gel (204 bp, 113 bp, and 155 bp), whereas, the Red deer PCR-RFLP fingerprints exhibited two fragments that appeared on the gel (268 bp and 204 bp), which were different from those of the Sika deer samples (Fig. 4b and 4c).
3.5 Cloning and sequencing of species fragments
Through the sequence comparison analysis of Sika deer and Red deer samples, it was found that there was a G/A SNP site on the Sika deer and Red deer data, in which the Sika deer was represented by G, and that of the Red deer by A, as was located on the Hpy188III restriction endonuclease recognition sequence (5′- TC^NNGA-3′), so that Sika-deer sequences were cut into three bands, and those of the Red deer were cut into two bands (Fig. 5a and 5b).
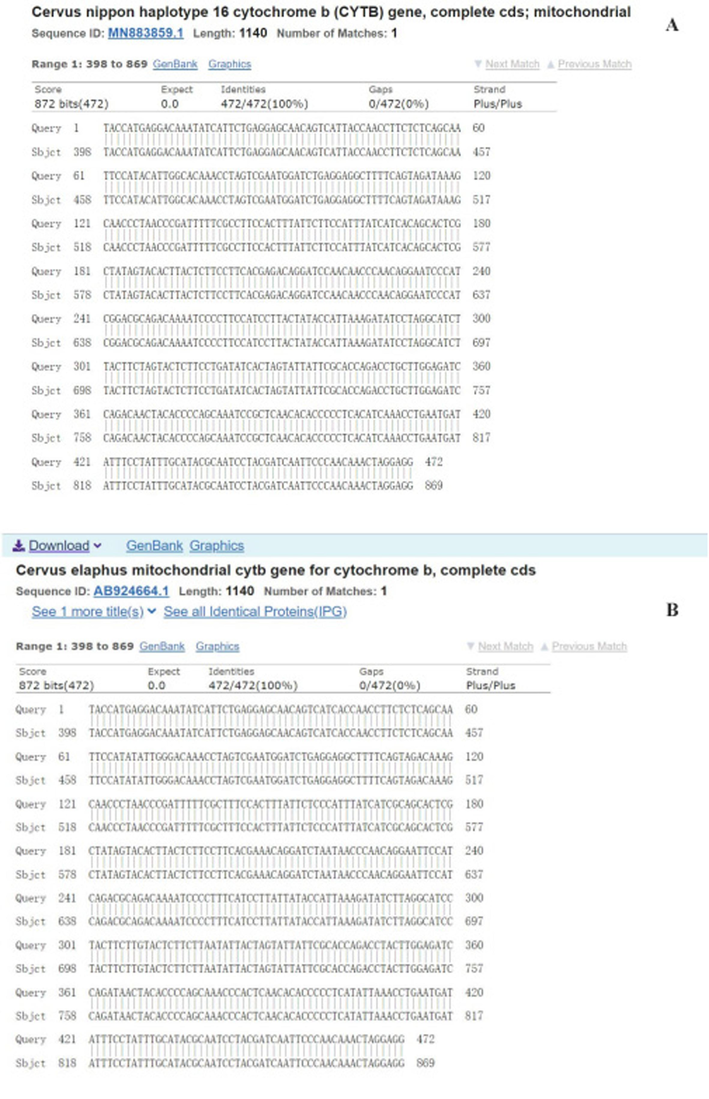
-
Comparison of partly Sika deer and Red deer Cyt b sequences A: C. nippon Cyt b sequences and blast with the Genbank B: C. elaphus Cyt b sequences and blast with the Genbank Footnote: Cyt b, cytochrome b.
3.6 Evaluation of the assay performance parameters
To validate the assay detection sensitivity, the Sika deer Penis et testis cervi DNA extracts were diluted with double-distilled water to 100 ng/μL. The PCR target band was detected at all dilutions except 0.0001 ng (Fig. 6a), showing that the minimum detection limit of the assay was determined to be 0.1 ng for deer Penis et testis cervi. To determine assay specificity, PCR amplification was performed with the mimic counterfeiting deer Penis et testis cervi samples containing material from both Sika deer and Red deer species. The reactions of templates with a ratio of 1:4 (weight/weight) showed clear patterns in the output diagram, PCR-RFLP findings indicated that no cross-contamination in any combination was observed, implying that the assay specificity was acceptable for authentication of deer Penis et testis cervi containing material from both Sika deer and Red deer species (Fig. 6b).
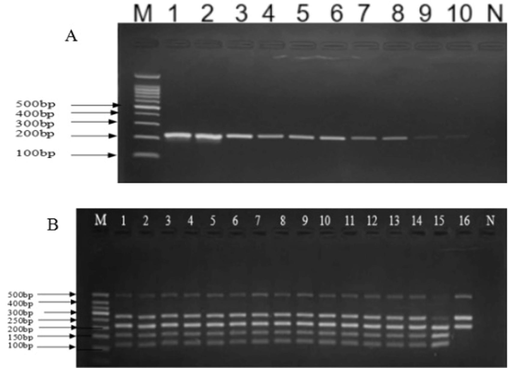
-
Agarose gel electrophoretogram of evaluation of the assay performance parameters A: Agarose gel electrophoretogram of the assay detection sensitivity Footnote: M−100 bp Ladder Marker;1. MHL-B(100 ng/μL);2. ML-B(100 ng/μL);3. MHL-B(10 ng/μL);4. ML-B(10 ng/μL);5. MHL-B(1.0 ng/μL);6. ML-B(1.0 ng/μL);7. MHL-B(0.1 ng/μL);8. ML-B(0.1 ng/μL);9. MHL-B(0.01 ng/μL);10. ML-B(0.001 ng/μL);N. Negative control MHL-B, Chinese spelling name (Mei hua lu bian), represents authentic Sika deer Penis et testis cervi (1. MHL2110008). ML-B, Chinese spelling name (Ma lu bian), represents authentic Red deer Penis et testis cervi (2. ML21101015). B: Agarose gel electrophoretogram of the assay detection specificity Footnote: M- 50 bp Ladder Marker;Mimic counterfeit deer Penis et testis cervi contained Sika deer and Red deer species, 1 ∼ 16 represents the ratio of 1:90, 1:80, 1:70, 1:60, 1:50, 1:40, 1:30, 1:20, 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, and 1:3 (weight/weight).
3.7 Identification of commercially available deer Penis et testis cervi samples
To validate the method, 30 batches of 60 commercially available deer Penis et testis cervi products were performed. Out of all sample, the former batches of 15 were labeled as Red deer species by sellers, S1-15, whereas, the latter batches of 15 were labeled as Sika deer species by sellers, S16-30. The results of species-specific PCR confirmed that all of which were authentic. However, The PCR-RFLP results found that sample S6 contained material from Sika deer, sample S16 contained material from Red deer species, which, although coincided with the results identified by the expert (Supplementary Table 1). The findings revealed a presence of mislabeling Red deer testis and penis as Sika deer on the markets. The result demonstrated that the accuracy of the two-step identification methods in this experiment was 100% (Fig. 7a to 7d).

-
Agarose gel electrophoretogram of evaluation of commercially available samples A: Agarose gel electrophoretogram of authenticity of commercially available Red deer Penis et testis cervi by a specific-species PCR Footnote: A:M- 100 bp Ladder Marker;1. S16;2. S17;3. S18;4. S19;5. S20;6. S21;7. S22;8. S23;9. S24;10. S25;11. S26;12. S27;13. S28;14. S29;15. S30;N. Negative control. B: Agarose gel electrophoretogram of authentic Red deer Penis et testis cervi by PCR-RFLP Footnote:M−50 bp Ladder Marker;1. S16;2. S17;3. S18;4. S19;5. S20;6. S21;7. S22;8. S23;9. S24;10. S25;11. S26;12. S28;13. S29;14. S30;N. Negative control C: Agarose gel electrophoretogram of authenticity of commercially available Sika deer Penis et testis cervi by a species PCR Footnote: M−100 bp Ladder Marker;1. S1;2. S2;3. S3;4. S4;5. S5;6. S6;7. S7;8. S8;9. S9;10. S10;11. S11;12. S12;13. S13;14. S14;15. S15;N. Negative control D: Agarose gel electrophoretogram of authentic Sika deer Penis et testis cervi by PCR-RFLP D:M- 50 bp Ladder Marker;1. S1;2. S2;3. S3;4. S4;5. S6;6. S7;7. S8;8. S10;9. S12;10. S14;11. S15;N. Negative control.
4 Discussion
There are many methods available with which to identify the authenticity of deer Penis et testis cervi, but there has been little research into the identification of sub-species. The traditional identification methods such as provenance and character required the appraiser to have extensive practical experience, and the identification results are often affected by subjectivity and are experience-dependent, with certain limitations. Although infraRed spectroscopy and high-performance liquid chromatography have high detection sensitivity, the routine maintenance of the instrument is cumbersome and the analysis is expensive, making is unsuitable for wider use (Ma et al.,2012; Lu et al.,2021).
In recent years, molecular biological identification technology has been popularized because of its advantages of accuracy, rapidity, and small sample size (Han et al.,2018). PCR-based assays were performed to identify the origin of the raw materials, especially in TCM (Bottero et al.,2003; Gao et al.,2016; Druml et al.,2015). Through gene sequence analysis, designed specific primers has become an important method for the identification of TCM materials of animal origin; however, it is difficult to identify TCM with relatively similar species (Verma and Singh, 2003). In comparison, PCR-RFLP fingerprints have several advantages over other DNA assay, such as a high level of reproducibility and ease of analysis in addition to optimal results from low-quality samples (Nesvadbová et al.,2019; Pfeiffer et al., 2004).
The origin of Sika deer Penis et testis cervi is similar to that from Red deer (Fan et al., 2021; Hoffmann et al.,2015), so an experiment was established using a combination of PCR and RFLP assays to realize differentiation between deer species. One of the keys of this study is the extraction of genomic DNA from all samples. Deer Penis et testis cervi products were mostly dried and fat-rich, thus affecting DNA extraction.
In this experiment, we managed to modify a sodium dodecyl sulfate (SDS) method supplemented with specific reagents to remove the fat and minerals. Finally, the modified SDS method was employed successfully to recovery the high quality genomic DNA from deer Penis et testis cervi samples. The concentration and purity of the genomic DNA obtained provided a good template for subsequent PCR amplification. So far, SDS, CTAB, and available commercial kits have been employed to extract and purify DNA from animal and plant materials. Of all methods, the SDS-based method has been reported to enable the highest DNA yield from animal matrix (Demekea et al., 2012; Stefanova et al., 2013). Xia et al, reported that the concentration of components in SDS lysis buffers varied widely. Moreover, the components supplemented with specific organic reagents to remove contaminants and precipitate DNA also vary between DNA isolation protocols (Xia et al., 2019). Optimization of the SDS-based method is a crucial step, which was sufficient to amplify the fragment in PCR-based technology. Our team have successfully developed a series of kits based on the modified SDS method used for the authenticity of deer fetus, mink heart, Zaocys dhumnades (Wang et al., 2019; Ai et al., 2017; Zhang et al., 2018). Besides that, a PCR kit for authentication of Panax quinquefolius L. with PCR-RFLP has been developed (Wang et al., 2022).
PCR-RFLP is an assay used to realize identification by cutting the specific target restriction site of DNA into segments of different lengths according to restriction endonuclease (Vaithiyanathan et al.,2021; Kumar et al., 2014). The selection of restriction endonuclease is an another key. We applied bioinformat-ics techniques to compare sequences and restriction endonuclease map analysis of Sika deer and Red deer’ Cyt b gene. Sika deer and Red deer’ PCR amp-licon has the different restriction enzyme sites for the Hpy188III. Therefore, we managed to establish a DNA fingerprinting with the PCR-RFLP method based on Cyt b gene region sequences of Sika deer and Red deer.Theoretically,the three distinct DNA fragments were observed in the Sika deer’ PCR ampli-con with two restriction enzyme sites for the Hpy188III and and two distinctDNA fragments were found in the Red deer PCR amplicon with a restriction enzyme site, respectively. Actually, the Sika deer characteristic PCR-RFLP fingerprints exhibited three distinct bands with the size of 204 bp,113 bp, and 155 bp, whereas, the Red deer PCR-RFLP fingerprints exhibited two bands that appeared on the gel with the size of 204 bp and 268 bp, which were different from those of the Sika deer samples (Fig. 4b and 4c).
Additionally, endonuclease concentration, digestion time and other factors may affect the accuracyof the experiment. This experiment was optimized to confirm that the concentration of Hpy188III endonuclease was between 10 and 40 U·μL−1, the digestion time was between 10 and 25 min. The results of repeated experiments confirmed that the combination of PCR and RFLP was reasonable and highly repeatable, which can accurately distinguish Sika deer Penis et testis cervi from that of the Red deer.
In the present study, the identification method established included two steps. The first step was to identify the authenticity of the genera of deer Penis et testis cervi, that is, it was necessary to determine whether the genera of deer Penis et testis was from Sika deer or Red deer. If it was genuine, one further step was required to determine the species. If it is a counterfeit product, the subsequent species identification is unnecessary. In the problem of TCM, the authenticity should be conducted first. After the authenticity is determined, the species identification of Sika deer Penis et testis cervi and Red deer Penis et testis cervi can be carried out according to the specific actual situation and clinical needs, which can meet the needs of practical sample detection.
There are some limitations to the present study. Our study was a preliminary study that needed confirmation on more size samples. Besides, the factors including different qualities of reagents and materials would influence the sensitivity and specificity on the test results, bringing about the potential for false positives or negatives. The standardized kit for the species identification of Penis et testis cervi could be developed to improve the stability and accuracy of the test results. These considerations will be validated in the future research.
5 Conclusion
A fast, accurate method for the identification of species of deer Penis et testis cervi was established. The method was simple and convenient, and showed strong applicability, overcoming the current limitations of intra-species identification of Sika deer Penis et testis cervi from those of the Red deer.
Acknowledgments
This study was supported by the Innovation Center for Detection Technology on the DNA Fingerprint of Traditional Chinese Medicine, Jilin Province, China.
Funds
This study received financial support from the Science and Technology Development Program of Jilin, China (20200404152YY and 20200403047SF). The Creative Funding Program of Chen Yu for graduate student (BH007).
Ethics approval
All animal samples collected complied with the Animal Research: Reporting In Vivo Experiments (ARRIVE) guidelines, and test protocols were approved by the Medicine Institutional Animal Care and Use Committee, Beihua University (number: 2021-1-22).
Consent to participate
All authors have consented to publication.
Consent to publication
All authors have approved to publish.
Code availability
Not applicable.
Authors' contributions
Tong Zhou conceived and designed the experiments and wrote a draft manuscript. Peiyao Li, Ying Zhang, Nan Wu, Yutong Zhang, Yangyang Li performed the experiments. Jinxia Ai and Mingcheng Li analyzed, interpreted the results of the experiments and revised the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- PCR-fingerprint profiles of mitochondrial and genomic DNA extracted from Fetus cervi using different extraction methods. Mitochondrial DNA A DNA Mapp Seq Anal.. 2017;28(6):781-786.
- [Google Scholar]
- Development of a PCR assay for the detection of animal tissues in ruminant feeds. J. Food Prot.. 2003;66:2307-2312.
- [Google Scholar]
- Pharmacopoeia of the People's Republic of China. Beijing: China Medical Science Press. Vol; 2010. p. :1.
- Mitochondrial DNA in wildlife forensic science: Species identification of tissues. Wildlife Soc Bull.. 1991;19:94-105.
- [Google Scholar]
- Demekea, T., Holigroski, M., Phan, A., 2012. Assessment of DNA extraction methods for PCR testing of discontinued or unapproved biotech events in single seeds of canola, flax and soybean. Food Control. 24, 44–49 10.1016.
- Authenticity control of game meat products–a single method to detect and quantify adulteration of fallow deer (Dama dama), Red deer (Cervus elaphus) and sika deer (Cervus nippon) by real-time PCR. Food Chem.. 2015;170:508-517.
- [Google Scholar]
- Development and validation of a rapid kit for authenticity of murine meat in meat products with a species-specific PCR assay. Food Additives & Contaminants: Part A.. 2020;37(5):1-9.
- [Google Scholar]
- Development and validation of a 1 K sika deer (Cervus nippon) SNP Chip. BMC Genom Data.. 2021;22(1):35.
- [Google Scholar]
- Development of multiplex PCR assay for authentication of Cornu Cervi Pantotrichum in traditional Chinese medicine based on cytochrome b and C oxidase subunit 1 genes. Mitochondrial DNA A DNA Mapp Seq Anal.. 2016;27(4):2989-2992.
- [Google Scholar]
- Application of Molecular Methods in the Identification of IngRedients in Chinese Herbal Medicines. Molecules. 2018;23:e2728.
- [Google Scholar]
- Species cross-amplification, identification and genetic variation of 17 species of deer (Cervidae) with microsatellite and mitochondrial DNA from antlers. Mol. Biol. Rep.. 2015;42(6):1059-1067.
- [Google Scholar]
- Authentication of beef, carabeef, chevon, mutton and pork by a PCR-RFLP assay of mitochondrial cytb gene. J. Food Sci. Technol.. 2014;51(11):3458-3463.
- [Google Scholar]
- Characteristics of PCR-SSCP and RAPD-HPCE methods for identifying authentication of Penis et testis cervi in Traditional Chinese Medicine based on cytochrome b gene. Mitochondrial DNA A DNA Mapp Seq Anal.. 2016;27(4):2757-2762.
- [Google Scholar]
- Simultaneous determination of 13 hormones in Testis et Penis Cervi using QuEChERS-ultra-high performance liquid chromatography-tandem mass spectrometry. Zhongguo Zhong Yao Za Zhi. 2021;46(24):6447-6453.
- [Google Scholar]
- Application of near infraRed spectroscopy in identi fication and quality control of traditional Chinese Medicin. J China Pharm.. 2012;23(7):661-663.
- [Google Scholar]
- PCR-RFLP identification of meat from Red deer, sika deer, roe deer, fallow deer, mouflon, wild boar, hare and cattle. Acta Vet. Brno. 2019;88(1):103-111.
- [Google Scholar]
- Establishment of a PCR Method for the Identification of Mink-Derived Components in Common Edible Meats. Journal of Analysis & Test.. 2021;001:006.
- [Google Scholar]
- Population genetic structure of Scombrops boops (Percoid, Scombropidae) around the Japanese archipelago inferRed from thecytochrome b gene sequence in mitochondrial DNA. Mitochondrial DNA. 2012;23:223-229.
- [Google Scholar]
- Diagnostic polymorphisms in the mitochondrial cytochrome b gene allow discrimination between cattle, sheep, goat, roe buck and deer by PCR-RFLP. BMC Genet.. 2004;5(5):30.
- [Google Scholar]
- A modified CTAB method for DNA extraction from soybean and meat products. Biotechnol. Biotec. Eq.. 2013;27:3803-3810.
- [Google Scholar]
- Research on the identification of penis et testis cervi with molecular taxonomy. Zhongguo Zhong Yao Za Zhi. 2002;27(8):573-575.
- [Google Scholar]
- Authentication of camel meat using species-specific PCR and PCR-RFLP. J. Food Sci. Technol.. 2021;58:3882-3889.
- [Google Scholar]
- Novel universal primers establish identity of an enormous number of animal species for forensic application. Mol. Ecol. Notes. 2003;3(1):28-31.
- [Google Scholar]
- Development and evaluation of a PCR Kit for authentication of Panax quinquefolius L. with PCR-Restriction Fragment Length Polymorphism. J. Anal. Test.. 2022;6:424-430.
- [Google Scholar]
- Protective effects of two kinds of aqueous extract of Penis et Testis Cervi against cisplatin-induced acute kidney injury in mice. Shanghai Journal of Traditional Chinese Medicine.. 2016;50(09):85-89.
- [Google Scholar]
- Comparison of anti-faigue effect and bioactive constituents in testis et Penis Cervi of Cervus nippon and Cervus elaphus. Food Sci. Technol. (. 2017;42(04):62-66.
- [Google Scholar]
- Establishment of mink heart identification method based on mitochondrial cytochrome b gene and development of its detection kit. Mitochondrial DNA A DNA Mapp Seq Anal.. 2019;30(2):325-331.
- [Google Scholar]
- Xia, Y., Chen. F., Du, Y., Liu, C., Bu, G., Xin, Y., Liu, B., 2019. A modified SDS-based DNA extraction method from raw soybean. Biosci Rep. 5; 39(2):BSR20182271.
- Molecular authentication of Chinese herbal materials. J. Food Drug Anal.. 2007;15:1.
- [Google Scholar]
- Development and evaluation of a PCR-based assay kit for authentication of Zaocys dhumnades in traditional Chinese medicine. Mitochondrial DNA A DNA Mapp Seq Anal.. 2018;29(1):102-106.
- [Google Scholar]
- Zhang, G.J., 2010. Research methodology of identification of traditional Chinese medicine. Beijing: People's Health Publishing House, 2010:1.
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.105337.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







