Translate this page into:
Development of a UHPLC-MS/MS-based data-mining method for rapid profiling and characterization of magnolol metabolites in rat urine and plasma
⁎Corresponding authors. wangzhibin4804@126.com (Zhibin Wang), crystal_wj@163.com (Jing Wang),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Magnolol, an essential bioactive natural lignans isolated from Magnolia Officinalis, has various pharmacological activities, such as antibacterial, anti-inflammatory, antioxidation and anti-tumor. In this study, a practical strategy based on ultra-high-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometry (UHPLC-LTQ-Orbitrap MS) was established to reveal the metabolic fate of magnolol in rat plasma and urine. First of all, in the data-dependent scanning (DDS) acquisition mode, the ESI-MSn data sets of biological samples and reference standard were obtained by using a high-quality online data analysis method. Subsequently, off-line data-mining techniques including multiple mass defect filters (MMDFs) and high-resolution extracted ion chromatograms (HREICs) were applied to screen major-to-trace metabolites of magnolol at the full-scan ESI-MS1 level. Finally, a total of 177 metabolites (prototype compound included) were preliminarily observed and characterized according to accurate mass measurement, the characteristic fragmentation patterns, chromato-graphic behaviors, and corresponding Clog P values. The results showed that magnolol experienced a variety of biotransformation reactions in rats, including hydroxylation, methoxylation, dehydrogenation, carboxylation, sulfonation, glucuronide conjugation, glucose conjugation, N-acetylcysteine (NAC) conjugation, glutathione conjugation and their composite reactions. In conclusion, this study not only greatly expanded the understanding of the therapeutic material basis and pharmacological mechanism of magnolol, but also provided new ideas for its toxicity evaluation, safety monitoring and drug delivery forms design.
Keywords
Magnolol
UHPLC-LTQ-Orbitrap MS
Data-mining techniques
Metabolites
1 Introduction
Magnolol (shown in Fig. 1), as a typical biphenylene lignans, is mainly extracted and isolated from magnolia plants, such as Magnolia officinalis Rehd. et Wils. and Magnolia officinalis (Rehd. et Wils.) Cheng subsp. biloba (Rehd.et Wils.) Law(Kawahara et al., 2020; Li et al., 2020; Santos et al., 2020; Zhang et al., 2020). In recent years, there has been increasing interest and research on the health benefits and extensive pharmacological properties of magnolol, such as antibacterial, anti-inflammatory, anti-tumor, antiviral, cardiovascular regulation and neuroprotection(Fukuyama et al., 2020; Kim et al., 2020; Kiseleva et al., 2020; Lovecká et al., 2020; Tao et al., 2020; Yuan et al., 2020). Therefore, it is used for the treatment and alleviation of various intractable diseases, including liver cancer, breast cancer, ischemic brain injury, chronic bronchitis and Alzheimer disease(Chen et al., 2020; Elhabak et al., 2020; Guo et al., 2020; Upadhyay et al., 2020; Xian et al., 2020). Moreover, as previously reported, magnolol could not only attenuate cisplatin-induced muscle wasting by M2c macrophage activation (Lee et al., 2020), but also prevent acute alcoholic liver damage by activating PI3K/Nrf2/PPARγ and inhibiting the NLRP3 signaling pathway (Liu et al., 2019). However, to our best knowledge, little is known until now about the biotransformation of magnolol in vivo, which is essential to explain and predict a variety of events related to the efficacy and toxicity of magnolol.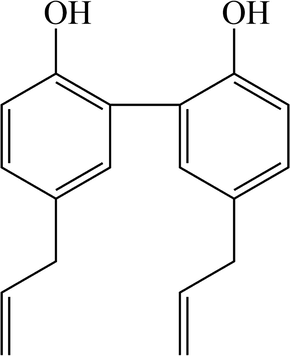
Structure information of magnolol.
In the past decade, ultra-high-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometry (UHPLC-LTQ-Orbitrap MS) has been broadly used in multi-component analysis and drug metabolism research of traditional Chinese medicine due to its advantages of high resolution, high quality accuracy and wide dynamic range (Stojković et al., 2020; Vasić et al., 2019; Zengin et al., 2020). By combining the full scan-parent ion list-dynamic exclusion (FS-PIL-DE) data acquisition method with multi-channel off-line LC-MS data-mining technology, it is possible to identify and capture trace metabolites overwhelmed by complex background noise or matrix interference. Along with the advancement of science and the shining of wisdom, multifarious data processing methods have emerged, including mass defect filter (MDF), multiple mass defect filters (MMDFs), high-resolution extracted ion chromatograms (HREICs), diagnostic product ions (DPIs), neutral loss fragments (NLFs), and background subtraction (BS), which further improve the accuracy and efficiency of metabolite identification (de Benzi, 2020; Lee et al., 2020; Shang et al., 2020; Stavrianidi, 2020; Xiao et al., 2020).
Herein, based on the combination of UHPLC-LTQ-Orbitrap mass spectrometer with multiple data-mining techniques, an integrated and effective strategy was established for the comprehensive identification and characterization of major-to-trace metabolites in plasma and urine of Sprague-Dawley (SD) rats after oral administration of magnolol. At the same time, this developed method would be further adopted to elucidate different biotransformation pathways of magnolol in rats.
2 Experiment
2.1 Chemicals and reagents
The reference standard of magnolol was purchased from Chengdu Must Biotechnology Co. Ltd (Sichuan, China). Its structure was fully elucidated by comparing its spectral data (ESI-MS and 1H, 13C NMR) with those published literature values. And its purity (≥98%, confirmed by HPLC-UV analysis) was suitable for UHPLC-HRMS analysis.
HPLC grade acetonitrile, methanol, and formic acid (FA) were all obtained from Thermo Fisher Scientific (Fair Lawn, NJ, USA). All other analytical grade chemicals were available from the workstation at the Beijing Chemical Works (Beijing, China). All deionized water used throughout the experiment was purified through the Milli-Q Gradient Å 10 system (Millipore, Billerica, MA, USA). Grace PureTM SPE C18-Low solid-phase extraction cartridges (200 mg/3 mL, 59 μm, 7 Å) were purchased from Grace Davison Discovery Science (Deerfield, IL, USA).
2.2 Animals
Eight male Sprague-Dawley (SD) rats weighing 220 g-240 g were purchased from Beijing Weitong Lihua Experimental Animals Company (Beijing, China). The animals were housed in a control room with standard temperature (24 ± 2℃) and humidity (70 ± 5%), and maintained in a 12-h light/12-h dark treatment. After a week of adaptation, the rats were randomly divided into two groups: Group A (n = 4), drug group for plasma and urine; Group B (n = 4), control group for blank plasma and urine. They were fasted for 12 h with free access to water prior to the experiment. The animal experimentation protocols were approved by the institutional Animal
Care and Use Committee at Beijing University of Chinese Medicine. The animal facilities and procedures were strictly in accordance with the Guide for the Care and Use of Laboratory Animals.
2.3 Drug administration and biological samples preparation
Magnolol was suspended in 0.5% carboxymethylcellulose sodium (CMC-Na) aqueous solution and given to Group A rats by gavage at a dose of 300 mg/kg body weight. Each rat in Group B was orally administered an equivalent amount of 0.5% CMC-Na aqueous solution. Blood samples (0.5 mL) were respectively collected from the suborbital venous plexus of rats at 0.5, 1.0, 1.5, 2.0, and 4.0 h after administration. After that, each sample was transferred to a heparinized micro-centrifuge tube and centrifuged at 3500 rpm (4℃) for 15 min to obtain plasma. Urine samples were collected over 0–24 h after oral administration. Finally, all biological samples from the same group were merged into a collective sample.
The solid-phase extraction (SPE) method, which could precipitate and concentrate proteins and solid residues, was applied to prepare all biological samples. Plasma and urine samples (1 mL) were separately added into the SPE cartridges, which were sequentially pretreated with methanol (5 mL) and deionized water (5 mL). Then, the SPE cartridges were washed with deionized water (5 mL) and methanol (3 mL) in turn. The methanol eluate was collected and evaporated to dryness under nitrogen at room temperature. The residue was re-dissolved in 100 μL of acetonitrile/water (10:90, v/v), and then centrifuged at 14000 rpm (4℃) for 15 min. The supernatant was used for further UHPLC-HRMS analysis.
2.4 Instrument and conditions
The LC analysis was carried out on the DIONEX Ultimate 3000 UHPLC system (Thermo Fisher Scientific, MA, USA), which was equipped with a binary pump, an automatic sampler and a column compartment. The chromatographic separation was performed at room temperature using a Waters ACQUITY BEH C18 column (2.1 × 100 mm i.d., 1.7 μm; Waters Corporation, Milford, MA, USA). Acetonitrile (solvent B) and 0.1% FA aqueous solution (solvent A) were used as the mobile phase. The flow rate was set to 0.30 mL/min and the linear gradient was as follows: 0–2 min, 5%-20% B; 2–27 min, 20%-85% B; 27–30 min, 85% B; 30–32 min, 85%-5% B; 32–35 min, 5% B. The injection volume was 2 μL.
High-resolution ESI-MS and MS/MS spectral analysis were performed on an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, MA, USA) that was connected to a UHPLC instrument via the ESI interface. Samples were analyzed in negative ion mode with the tuning method set as follows: sheath gas (nitrogen) flow rate of 40 arb, auxiliary gas (nitrogen) flow rate of 20 arb, capillary temperature of 350 °C, electrospray voltage of 4.0 kV, capillary voltage of 35 V, and tube lens voltage of 110 V. Centroided mass spectra was obtained within the mass range of m/z 50–800.
In the full-scan (FS) experiment, the HRMS data was recorded at mass resolving power of 30,000 full width at half maximum (FWHM, calculated for m/z 200). To minimize the total analysis time, the data-dependent MS/MS scanning was performed to trigger the fragmentation mass spectra of the target ions. The collision energy of the collision-induced dissociation (CID) was adjusted to 40% of the maximum value. The function of dynamic exclusion to prevent repetition was enabled, and the repeat count was set to 5 with the dynamic repeat time at 30 s, and the dynamic exclusion duration was 60 s. In addition, the parent ion list-dynamic exclusion (PIL-DE) dependent acquisition mode was also used as a complementary method to obtain the ESI-MSn stages of the screened metabolite candidates. Data-dependent ESI-MS2 analyses were triggered by the three most-abundant ions in the list of predicted metabolite precursors while ESI-MS3 analyses of the most-abundant product ions were followed.
2.5 Peak selections and data processing
The Thermo Xcalibur 2.1 workstation was applied to collect and process the HR-ESI-MS1 and MSn data. In order to obtain as many ESI-MS/MS fragment ions of magnolol metabolites as possible, the peaks detected with intensity over 10,000 were selected for further structural identification. The chemical formulas for all parent ions of the selected peaks were calculated from the accurate mass using a formula predictor by setting the parameters as follows: C [0–30], H [0–50], O [0–20], S [0–4], N [0–3] and ring double bond (RDB) equivalent value [0–15]. Other elements, such as P and Br, were not considered because they rarely appeared in the complex matrix.
3 Results and discussion
3.1 The establishment of analytical strategy
In this study, based on UHPLC-LTQ-Orbitrap MS analysis combined with data acquisition and various post-acquisition data processing techniques, a systematic and effective strategy (shown in Fig. 2) was proposed for the rapid screening and characterization of magnolol metabolites in rats. First of all, high-quality ESI-MS1 analysis was performed and ESI-MSn datasets for biological samples and reference standard were obtained in data-dependent scanning (DDS) acquisition mode. Secondly, for the subsequent post-acquisition data-mining processing, a combination of HREICs and MMDFs method was adopted for the comprehensive screening of magnolol metabolites. Among them, HREICs were utilized to identify known (as previously reported in literatures) and predicted metabolites (based on common biotransformation reactions), while MMDFs were employed to acquire specific HR-MS1 information for unknown and unpredictable metabolites. Then, the ESI-MSn datasets of all screened metabolite candidates were collected by the full scan-parent ion list-dynamic exclusion (FS-PIL-DE) data acquisition method. Afterwards, according to the chromatographic retention times, accurate mass measurements, characteristic mass fragmentation behaviors and the corresponding Clog P values, the structures of magnolol metabolites were tentatively expounded. Finally, the metabolic pathways of magnolol in rats were positively summarized based on the identified metabolites and the corresponding biotransformation reactions.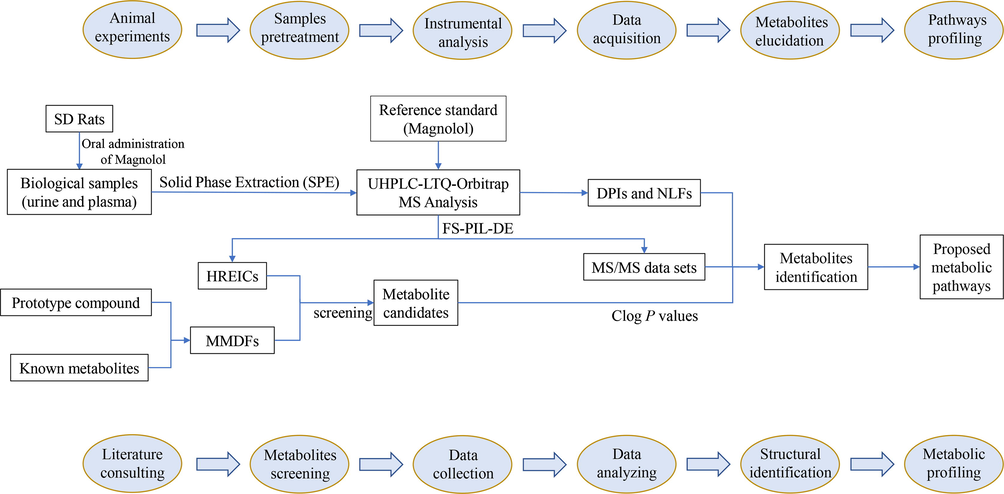
Summary diagram of the developed analytical strategy and methodology.
3.2 Fragmentation behaviors of the magnolol in negative ion mode
In order to better understand the characteristic fragmentation behaviors of magnolol, a comprehensive analysis of the reference standard solution was implemented using UHPLC-LTQ-Orbitrap MS to obtain the unique DPIs and NLFs of magnolol. In the negative ion mode, magnolol showed [M−H]- ion at m/z 265.12357 (C18H17O2, 4.77 ppm) in the ESI-MS1 spectrum. In its ESI-MS2 spectrum, several diagnostic product ions at m/z 247, m/z 237, m/z 224 and m/z 197 were generated due to the loss of H2O (18 Da), CO (28 Da), C3H5 (41 Da) and C3H5 + C2H3 (68 Da), respectively. Besides, the NLFs of 2 Da (m/z 247 → m/z 245 → m/z 243) and 26 Da (m/z 247 → m/z 221) were both observed in the ESI-MS3 spectrum, which further illustrated the fragmentation patterns of m/z 247. If different kinds of biotransformation reactions occurred on the original drug, the new DPIs at m/z 247 + X, m/z 237 + X, and m/z 224 + X (X = molecular weight of substituent groups, such as 14 (CH2), 30 (OCH2), and 162 (Glc)) derived from the newly produced compounds would provide an important basis for the structural identification of the metabolites. The proposed fragmentation pathways of magnolol were illustrated in Fig. 3, and the ESI-MSn spectra of magnolol in negative ion mode were displayed in Fig. 4.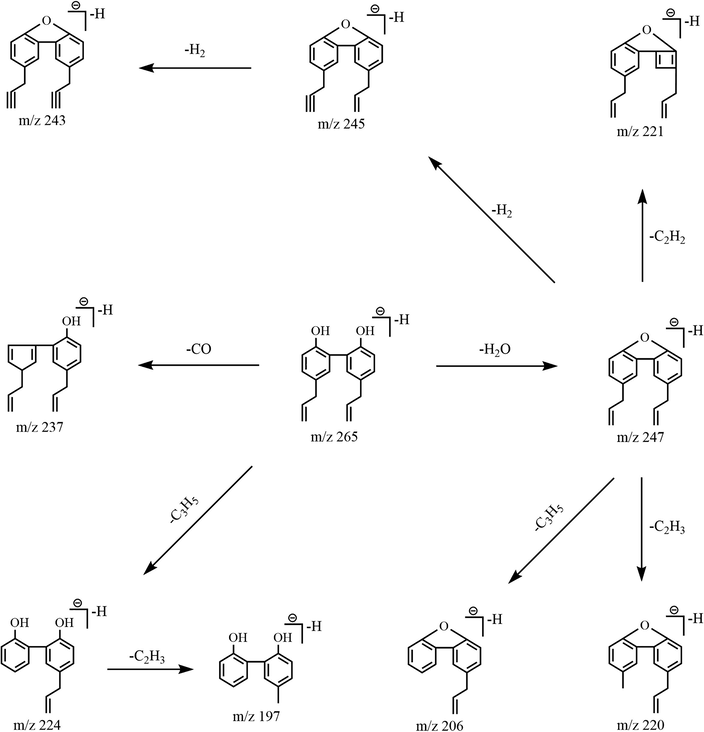
The mass fragmentation behaviors of magnolol.
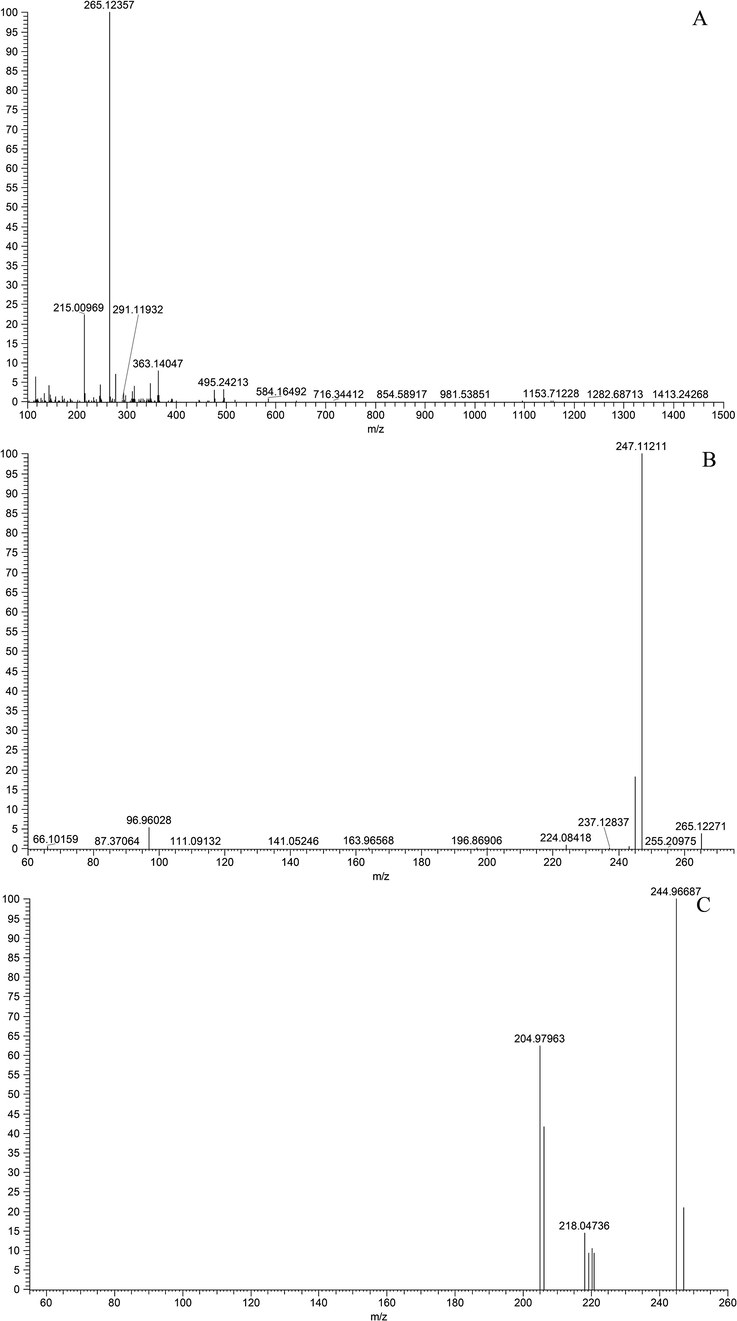
ESI-MSn spectra of magnolol in negative ion mode: (A) MS1 spectrum; (B) MS2 spectrum; (C) MS3 spectrum.
3.3 Implement of MMDFs data-mining methods
In order to reduce the potential interferences of endogenous substances, the MMDFs approach was chosen as a crucial complement to the HREICs method to entirely obtain the HR-MS1 datasets of low-level predicted and unpredictable metabolites. For the MMDFs method, the most critical step was to set up metabolite templates, which commonly included drug filter, substructure filter, and conjugate filter (Zhang et al., 2015). Since magnolol is difficult to generate two small molecules through the cleavage reaction, three templates were founded in parallel to screen relevant compounds:1) parent drug template (m/z 266.13013) and its conjugation templates (m/z 346.08695 for sulfate conjugation, m/z 442.16222 for glucuronide conjugation); 2) hydroxylated magnolol template (m/z 282.12504) and its conjugation templates (m/z 362.08186 for sulfate conjugation, m/z 458.15713 for glucuronide conjugation);3) carboxylated magnolol template (m/z 296.10431) and its conjugation templates (m/z 376.06113 for sulfate conjugation, m/z 472.13640 for glucuronide conjugation). According to the above-mentioned metabolites screening templates, each MDF window was set to ± 50 mDa around the mass defects of the templates over a mass range of ± 50 Da around the filter template masses. Based on the similarity of mass defects of metabolites and their core substructures, the MMDFs method could mine major-to-trace metabolites to the greatest extent from complex background noise and endogenous components.
3.4 Identification of magnolol metabolites
A total of 177 metabolites were detected and characterized in rat urine and plasma using UHPLC-LTQ-Orbitrap MS technology in combination with the established strategy. Among them, 166 metabolites were found in the urine, while 26 metabolites were derived from plasma. The chromatographic and mass spectral data for all metabolites were presented in Table 1, and their corresponding HREICs were shown in Fig. 5. Note: tR: retention time; U: urine; P: plasma; +: detected; −: undetected.
Peak
tR/min
Formula [M + H]+
Theoretical Mass m/z
Experimental Mass m/z
Error
(ppm)MS/MS fragment ions
Identification/Reactions
U
P
M0
17.34
C18H17O2
265.12229
265.12357
4.77
MS2[2 6 5]:265(1 0 0),247(60),245(11),224(1)
MS3[2 4 7]:206(1 0 0),245(38),203(15),247(14)Magnolol
+
+
M1
2.92
C18H21O6
333.13319
333.13571
7.34
MS2[3 3 3]:273(1 0 0),213(32),226(3)
Loss of 2C2H2, tetramethoxylation
+
−
M2
3.39
C18H21O6
333.13319
333.13589
7.88
MS2[3 3 3]:273(1 0 0),213(22),315(4),281(3)
Loss of 2C2H2, tetramethoxylation
+
−
M3
3.49
C21H19O9
415.10249
415.10593
8.60
MS2[4 1 5]:295(1 0 0),267(8),325(2),200(1)
MS3[2 9 5]:267(1 0 0),253(1)Loss of C3H4, oxidation, dehydrogenation, dihydroxylation, glucose conjugation
+
−
M4
3.67
C18H19O8S
395.07949
395.08267
7.99
MS2[3 9 5]:315(1 0 0),339(6),377(2)
MS3[3 1 5]:297(1 0 0),237(49),255(7),267(4)Loss of C2H2, dimethoxylation, hydroxylation, sulfonation
+
−
M5
3.77
C18H19O5
315.12269
315.12534
8.38
MS2[3 1 5]:297(1 0 0),237(63),255(6),267(2),272(1)
MS3[2 9 7]:237(1 0 0),219(4),222(2)Loss of C2H2, dimethoxylation, hydroxylation
+
−
M6
3.87
C15H9O4
253.04971
253.05196
9.58
MS2[2 5 3]:253(1 0 0),224(55),209(40),225(34),197(20)
Loss of C3H4, oxidation, dehydrogenation,
dihydroxylation+
−
M7
3.87
C18H17O5
313.10701
313.10971
8.50
MS2[3 1 3]:295(1 0 0),237(26),253(8),235(2)
MS3[2 9 5]:237(1 0 0),265(64),235(56),223(42),267(12)Loss of 2CH2, dimethoxylation, hydroxylation
+
−
M8
3.87
C21H19O9
415.10249
415.10587
8.46
MS2[4 1 5]:253(1 0 0),252(15),295(6),315(3),229(2)
MS3[2 5 3]:224(1 0 0),225(30)Loss of C3H4, oxidation, dehydrogenation, dihydroxylation, glucose conjugation
+
−
M9
3.87
C21H17O10
429.08179
429.08514
8.20
MS2[4 2 9]:253(1 0 0),175(39),411(6),349(5)
Loss of C3H4, oxidation, dehydrogenation, dihydroxylation, glucuronide conjugation
+
−
M10
3.97
C16H11O5
283.06019
283.06284
9.68
MS2[2 8 3]:268(1 0 0),255(2),220(1)
MS3[2 6 8]:240(1 0 0),224(1)Loss of C2H2, oxidation, carboxylation, hydroxylation
+
−
M11
4.08
C16H15O5
287.09149
287.09375
8.19
MS2[2 8 7]:227(1 0 0),209(9),269(8),209(6),219(2),237(1)
MS3[2 2 7]:209(1 0 0),181(22)Loss of C3H4 + CH2, dimethoxylation,
hydroxylation+
−
M12
4.28
C18H19O5
315.12269
315.12555
9.04
MS2[3 1 5]:297(1 0 0),237(81),255(5),267(4)
MS3[2 9 7]:237(1 0 0),219(3),270(2)Loss of C2H2, dimethoxylation, hydroxylation
+
−
M13
4.28
C21H19O9
415.10249
415.10596
8.68
MS2[4 1 5]:253(1 0 0),252(70),295(6),335(5),292(4)
Loss of C3H4, oxidation, dehydrogenation, dihydroxylation, glucose conjugation
+
−
M14
4.48
C24H25O10
473.14422
473.14786
7.69
MS2[4 7 3]:297(1 0 0),436(5),427(4),269(3),275(2)
MS3[2 9 7]:279(1 0 0),250(11),261(7)Dihydroxylation, glucuronide conjugation
+
−
M15
4.57
C15H9O5
269.04459
269.04712
9.92
MS2[2 6 9]:269(1 0 0),181(48),225(44),201(38),241(21)
Loss of C3H4, oxidation, carboxylation, hydroxylation
+
−
M16
4.57
C18H19O5
315.12269
315.12524
8.06
MS2[3 1 5]:267(1 0 0),297(4),255(2),224(1)
MS3[2 6 7]:249(1 0 0),247(29),239(28),221(10)Loss of C2H2, dimethoxylation, hydroxylation
+
−
M17
4.57
C18H19O8S
395.07949
395.08282
8.37
MS2[3 9 5]:315(1 0 0),267(12)
MS3[3 1 5]:267(1 0 0),297(5)Loss of C2H2, dimethoxylation, hydroxylation, sulfonation
+
−
M18
4.57
C15H11O4
255.06532
255.06772
9.94
MS2[2 5 5]:225(1 0 0),214(33),211(25),240(10)
Loss of C3H4 + CH2, dehydrogenation,
methoxylation, hydroxylation+
−
M19
4.68
C18H17O5
313.10701
313.10983
8.88
MS2[3 1 3]:253(1 0 0),295(23),235(7),207(6)
MS3[2 5 3]:235(1 0 0),207(84),225(10)Loss of 2CH2, dimethoxylation, hydroxylation
+
−
M20
4.88
C15H9O4
253.04971
253.05193
9.46
MS2[2 5 3]:253(1 0 0),209(61),224(58),225(50),197(22)
Loss of C3H4, oxidation, dehydrogenation, dihydroxylation
+
−
M21
4.88
C15H9O7S
333.00652
333.00919
8.53
MS2[3 3 3]:253(1 0 0),200(5),232(1)
MS3[2 5 3]:211(1 0 0),209(36),225(29),197(11)Loss of C3H4, oxidation, dehydrogenation, dihydroxylation, sulfonation
+
−
M22
5.00
C16H11O5
283.06019
283.06271
9.22
MS2[2 8 3]:268(1 0 0),66(2),255(1)
MS3[2 6 8]:240(1 0 0),239(4),224(1)Loss of C2H2, oxidation, carboxylation, hydroxylation
+
−
M23
5.00
C18H17O6
329.10189
329.10480
8.62
MS2[3 2 9]:137(1 0 0),311(31),219(23),269(6),226(5)
Tetrahydroxylation
+
−
M24
5.00
C29H33O16
637.17634
637.18103
7.41
MS2[6 3 7]:461(1 0 0),619(1)
MS3[4 6 1]:239(1 0 0),299(85),281(80),263(13)Loss of C2H2, dimethoxylation, glucuronide conjugation, glucose conjugation
+
−
M25
5.10
C24H29O9
461.18059
461.18439
8.20
MS2[4 6 1]:239(1 0 0),299(93),281(78)
Loss of C2H2, dimethoxylation,
glucose conjugation+
−
M26
5.20
C15H9O5
269.04459
269.04700
9.48
MS2[2 6 9]:241(1 0 0),213(29),225(10),253(7)
MS3[2 4 1]:213(1 0 0),197(8)Loss of C3H4, oxidation, carboxylation, hydroxylation
+
−
M27
5.43
C24H27O10
475.15989
475.16415
9.00
MS2[4 7 5]:299(1 0 0),175(3),239(1)
MS3[2 9 9]:239(1 0 0),281(7),221(4)Loss of C2H2, dimethoxylation,
glucuronide conjugation+
−
M28
5.54
C15H9O5
269.04459
269.04712
9.92
MS2[2 6 9]:269(1 0 0),225(46),181(41),241(26),197(18)
Loss of C3H4, oxidation, carboxylation, hydroxylation
+
−
M29
5.54
C24H29O9
461.18059
461.18381
6.94
MS2[4 6 1]:299(1 0 0),239(23),267(12)
Loss of C2H2, dimethoxylation,
glucose conjugation+
−
M30
5.63
C18H19O5
315.12269
315.12534
8.38
MS2[3 1 5]:255(1 0 0),200(11),267(10),214(9)
Loss of C2H2, dimethoxylation, hydroxylation
+
−
M31
5.73
C21H22O5NS
400.12132
400.12466
8.35
MS2[4 0 0]:279(1 0 0),341(26),200(14),333(10)
Hydroxylation, cysteine conjugation
+
−
M32
5.82
C18H19O4
299.12771
299.13010
7.74
MS2[2 9 9]:239(1 0 0),281(3),221(2),133(1),263(1)
MS3[2 3 9]:221(1 0 0),219(18),133(13),198(2),209(1)Loss of C2H2, dimethoxylation
+
+
M33
5.82
C18H19O7S
379.08459
379.08713
6.67
MS2[3 7 9]:299(1 0 0),222(2),281(1)
MS3[2 9 9]:239(1 0 0),221(2),281(2),225(1)Loss of C2H2, dimethoxylation, sulfonation
+
+
M34
5.95
C18H13O2
261.09107
261.09323
8.52
MS2[2 6 1]:233(1 0 0),205(15),231(7),203(6)
Oxidation, dehydrogenation
+
−
M35
6.06
C24H25O10
473.14422
473.14767
7.29
MS2[4 7 3]:297(1 0 0),201(7),239(6),235(5)
Dihydroxylation, glucuronide conjugation
+
−
M36
6.18
C18H19O5
315.12269
315.12521
7.97
MS2[3 1 5]:267(1 0 0),297(6),255(5),237(1)
MS3[2 6 7]:249(1 0 0),239(23),221(8),224(1)Loss of C2H2, dimethoxylation, hydroxylation
+
−
M37
6.26
C18H19O4
299.12771
299.13013
7.84
MS2[2 9 9]:239(1 0 0),281(3),221(2),133(1),263(1)
MS3[2 3 9]:221(1 0 0),219(14),133(9),237(4)Loss of C2H2, dimethoxylation
+
+
M38
6.26
C18H19O7S
379.08459
379.08719
6.83
MS2[3 7 9]:299(1 0 0),297(1),242(1)
MS3[2 9 9]:239(1 0 0),281(3),221(2),263(1)Loss of C2H2, dimethoxylation, sulfonation
−
+
M39
6.29
C14H9O2
209.05991
209.06119
7.10
MS2[2 0 9]:66(1 0 0),209(82),181(74),208(41),163(30)
Loss of C3H4 + CH2, dehydrogenation
+
−
M40
6.29
C14H9O3
225.05481
225.05592
5.77
MS2[2 2 5]:207(1 0 0),66(84),200(30),157(29),189(23)
Loss of C3H4 + CH2, dehydrogenation,
Hydroxylation+
−
M41
6.29
C15H9O4
253.04971
253.05106
6.03
MS2[2 5 3]:253(1 0 0),224(3),209(3),225(2),197(1)
Loss of C3H4, oxidation, dehydrogenation, dihydroxylation
+
−
M42
6.29
C17H15O3
267.10151
267.10275
4.42
MS2[2 6 7]:249(1 0 0),239(38),133(13),221(13),247(4)
MS3[2 4 9]:221(1 0 0),231(58),247(23),195(12)Loss of CH2, hydroxylation
+
−
M43
6.40
C19H21O5
329.13829
329.14038
6.17
MS2[3 2 9]:299(1 0 0),297(6),237(65),269(41),239(11)
Loss of C2H2, trimethoxylation
+
−
M44
6.40
C24H27O10
475.15989
475.16324
7.09
MS2[4 7 5]:299(1 0 0),175(10)
MS3[2 9 9]:239(1 0 0),281(5),221(2)Loss of C2H2, dimethoxylation,
glucuronide conjugation+
−
M45
6.40
C15H11O4
255.06532
255.06677
6.21
MS2[2 5 5]:254(1 0 0),237(4),225(3),209(1)
MS3[2 5 4]:224(1 0 0),192(17),210(14)Loss of C3H4 + CH2, dehydrogenation,
methoxylation, hydroxylation+
−
M46
6.40
C19H19O6
343.08342
343.08679
6.57
MS2[3 4 3]:229(1 0 0),283(44),325(43),317(25),328(24)
Loss of CH2, dimethoxylation, dihydroxylation
+
−
M47
6.52
C16H15O3
255.10159
255.10394
9.29
MS2[2 5 5]:254(1 0 0),237(5),210(4),225(3),226(2)
MS3[2 5 4]:224(1 0 0),192(17),210(14),225(13)Loss of C2H2, hydroxylation
+
−
M48
6.62
C15H9O5
269.04459
269.04208
−8.81
MS2[2 6 9]:240(1 0 0),241(17),225(4),197(1)
Loss of C3H4, oxidation, carboxylation, hydroxylation
+
−
M49
6.62
C16H11O5
283.06019
283.06235
7.95
MS2[2 8 3]:268(1 0 0),255(1)
MS3[2 6 8]:240(1 0 0),239(5),224(3)Loss of C2H2, oxidation, carboxylation, hydroxylation
+
−
M50
6.62
C15H13O4
257.08099
257.08304
8.58
MS2[2 5 7]:135(1 0 0),239(3),215(2),242(1)
MS3[1 3 5]:91(1 0 0)Loss of C2H2 + CH2, dihydroxylation
+
−
M51
6.73
C18H17O4
297.11211
297.11447
7.86
MS2[2 9 7]:237(1 0 0),267(86),225(75),279(19),261(11)
Loss of 2CH2, dimethoxylation
+
−
M52
6.73
C24H23O10
471.12859
471.13153
6.28
MS2[4 7 1]:295(1 0 0),382(3),221(4)
MS3[2 9 5]:251(1 0 0),233(13),277(3)Carboxylation, glucuronide conjugation
+
−
M53
6.73
C18H21O4
301.14349
301.14542
6.59
MS2[3 0 1]:283(1 0 0),240(82),258(19),273(14)
Hydrogenation, dihydroxylation
+
−
M54
6.73
C19H21O5
329.13829
329.14059
6.81
MS2[3 2 9]:269(1 0 0),311(16),267(2),299(1)
MS3[2 6 9]:251(1 0 0),241(44),233(15)Loss of C2H2, trimethoxylation
+
−
M55
6.73
C18H17O7S
377.06892
377.07141
6.52
MS2[3 7 7]:297(1 0 0),263(2),341(1)
MS3[2 9 7]:237(1 0 0),225(82),267(58),253(31)Loss of 2CH2, dimethoxylation, sulfonation
+
−
M56
6.84
C16H15O5
287.09149
287.09399
9.02
MS2[2 8 7]:227(1 0 0),66(92),200(37),192(22)
Loss of C3H4 + CH2, dimethoxylation,
hydroxylation+
−
M57
6.84
C24H27O10
475.15989
475.16385
8.37
MS2[4 7 5]:299(1 0 0),457(13),281(7),175(4)
MS3[2 9 9]:239(1 0 0),221(6),281(3)Loss of C2H2, dimethoxylation,
glucuronide conjugation+
−
M58
6.84
C24H25O10
473.14422
473.14822
8.45
MS2[4 7 3]:297(1 0 0),267(3),383(2),307(2)
MS3[2 9 7]:267(1 0 0),279(51),237(19)Loss of 2CH2, dimethoxylation,
glucuronide conjugation+
−
M59
6.95
C24H29O9
461.18059
461.18427
7.94
MS2[4 6 1]:299(1 0 0),239(87),281(50),263(11)
MS3[2 9 9]:239(1 0 0)Loss of C2H2, dimethoxylation,
glucose conjugation+
−
M60
7.05
C17H15O2
251.10661
251.10907
9.61
MS2[2 5 1]:233(1 0 0),231(10),66(2),133(1)
MS3[2 3 3]:231(1 0 0),206(45)Loss of CH2
+
−
M61
7.05
C18H17O3
281.11721
281.11996
9.74
MS2[2 8 1]:263(1 0 0),245(43),133(7),233(4),261(2)
MS3[2 6 3]:245(1 0 0),243(5),235(4),133(3)Hydroxylation
+
−
M62
7.05
C18H17O5
313.10701
313.10928
7.12
MS2[3 1 3]:298(1 0 0),285(2),269(1)
MS3[2 9 8]:283(1 0 0),270(51),255(28)Loss of 2CH2, dimethoxylation, hydroxylation
+
−
M63
7.05
C17H13O6
313.07059
313.07333
8.51
MS2[3 1 3]:298(1 0 0),285(3),191(2),269(1),253(1)
MS3[2 9 8]:283(1 0 0),270(54),255(16),239(3),268(2)Loss of C2H2, oxidation, carboxylation, hydroxylation, methoxylation
+
−
M64
7.17
C21H22O5NS
400.12132
400.12454
8.05
MS2[4 0 0]:279(1 0 0),216(8),200(7),267(6)
Hydroxylation, cysteine conjugation
+
−
M65
7.38
C17H17O3
269.11729
269.11871
5.53
MS2[2 6 9]:66(1 0 0),251(90),239(71),225(52),221(29)
Loss of C2H2, methoxylation
+
−
M66
7.38
C24H25O10
473.14422
473.14774
7.43
MS2[4 7 3]:297(1 0 0),200(7),222(5),351(4)
Dihydroxylation, glucuronide conjugation
+
−
M67
7.49
C17H15O2
251.10661
251.10878
8.46
MS2[2 5 1]:233(1 0 0),231(9),210(1),183(1)
MS3[2 3 3]:231(1 0 0),206(7),189(3)Loss of CH2
+
−
M68
7.49
C18H17O3
281.11721
281.11963
8.57
MS2[2 8 1]:263(1 0 0),245(41),133(7),233(4),261(2)
MS3[2 6 3]:245(1 0 0),133(13),243(5),235(4)Hydroxylation
+
−
M69
7.49
C14H11O4
243.06539
243.06720
8.29
MS2[2 4 3]:66(1 0 0),175(79),214(66),109(65),225(43),181(28)
Loss of C3H4 + CH2, dihydroxylation
+
−
M70
7.60
C18H15O2
263.10661
263.10895
8.72
MS2[2 6 3]:245(1 0 0),133(28),235(4),243(2)
MS3[2 4 5]:245(1 0 0),217(94),243(35),204(10)Dehydrogenation
+
−
M71
7.60
C15H11O5
271.06029
271.06250
8.85
MS2[2 7 1]:165(1 0 0),151(5),66(2),137(1),227(1)
MS3[1 6 5]:137(1 0 0),121(28),109(8),93(7)Carboxylation, loss of C3H4, hydroxylation
+
−
M72
7.60
C18H17O4S
329.08422
329.08661
7.30
MS2[3 2 9]:249(1 0 0),208(16),222(2)
MS3[2 4 9]:235(1 0 0),221(44),208(15)Dehydroxylation, sulfonation
+
−
M73
7.72
C16H13O2
237.09111
237.09323
9.38
MS2[2 3 7]:209(1 0 0),196(37),222(14),235(5),193(4),219(2)
MS3[2 0 9]:168(1 0 0),194(87),207(57),181(27)Loss of C2H2, dehydrogenation
+
−
M74
7.72
C17H13O3
265.08581
265.08841
9.39
MS2[2 6 5]:237(1 0 0),209(13),196(10),247(6),221(2)
MS3[2 3 7]:209(1 0 0),196(57),235(9),193(4)Loss of CH2, dehydrogenation, hydroxylation
+
−
M75
7.72
C17H15O4
283.09641
283.09906
9.09
MS2[2 8 3]:265(1 0 0),225(15),237(7),239(2),221(1)
MS3[2 6 5]:237(1 0 0),209(10),247(9),221(5)Loss of CH2, dihydroxylation
+
−
M76
7.72
C18H17O5
313.10701
313.10971
8.50
MS2[3 1 3]:283(1 0 0),265(3),295(1)
MS3[2 8 3]:265(1 0 0),225(64),237(4)Loss of 2CH2, dimethoxylation, hydroxylation
+
−
M77
7.72
C18H17O7S
377.06892
377.07181
7.59
MS2[3 7 7]:297(1 0 0),315(51),271(14),359(10)
Dihydroxylation, sulfonation
+
−
M78
7.83
C24H25O9
457.14931
457.15280
7.64
MS2[4 5 7]:281(1 0 0),439(11),175(3)
MS3[2 8 1]:263(1 0 0),245(34),133(8),233(4)Hydroxylation, glucuronide conjugation
+
−
M79
7.95
C18H17O3
281.11721
281.11850
4.55
MS2[2 8 1]:263(1 0 0),245(22),133(7),253(2),235(1)
MS3[2 6 3]:245(1 0 0),133(32),243(5),235(4)Hydroxylation
+
+
M80
7.95
C18H17O6S
361.07401
361.07550
4.06
MS2[3 6 1]:281(1 0 0),331(4),333(2)
MS3[2 8 1]:263(1 0 0),245(17),133(5)Hydroxylation, sulfonation
+
−
M81
7.95
C24H23O10
471.12859
471.13135
5.89
MS2[4 7 1]:453(1 0 0),427(90),295(76),361(23)
Carboxylation, glucuronide conjugation
+
−
M82
7.95
C17H17O3
269.11729
269.11807
3.16
MS2[2 6 9]:269(1 0 0),240(4),225(3),201(3),197(2),241(1)
Loss of C2H2, methoxylation
+
−
M83
7.95
C24H25O10
473.14422
473.14642
4.64
MS2[4 7 3]:297(1 0 0),175(13),295(7),417(4)
Dihydroxylation, glucuronide conjugation
+
−
M84
8.06
C14H9O3
225.05481
225.05600
6.13
MS2[2 2 5]:181(1 0 0),197(99),169(26),207(7)
Loss of C3H4 + CH2, dehydrogenation,
Hydroxylation+
−
M85
8.06
C24H25O9
457.14931
457.15140
4.58
MS2[4 5 7]:281(1 0 0),295(23),439(7),175(4)
MS3[2 8 1]:263(1 0 0),245(23),133(14),235(3)Hydroxylation, glucuronide conjugation
+
−
M86
8.06
C15H9O5
269.04459
269.04614
6.28
MS2[2 6 9]:269(1 0 0),181(4),241(3),225(2),197(1)
Loss of C3H4, oxidation, carboxylation, hydroxylation
+
−
M87
8.06
C15H11O5
271.06029
271.05988
−0.81
MS2[2 7 1]:270(1 0 0),151(12),182(6),226(5),202(4),242(2)
MS3[2 7 0]:270(1 0 0),226(40),198(21),224(17)Carboxylation, loss of C3H4, hydroxylation
+
−
M88
8.06
C16H13O4
269.08092
269.07828
−9.50
MS2[2 6 9]:269(1 0 0),181(4),225(3),197(2)
Loss of 2CH2, dihydroxylation
+
−
M89
8.17
C18H17O6S
361.07401
361.07623
6.08
MS2[3 6 1]:281(1 0 0),305(10),333(3),331(1)
MS3[2 8 1]:263(1 0 0),245(11),133(5),233(1)Hydroxylation, sulfonation
+
−
M90
8.17
C24H23O10
471.12859
471.13153
6.28
MS2[4 7 1]:295(1 0 0),277(44),453(18),361(11)
MS3[2 9 5]:251(1 0 0),231(3),277(2),233(1)Carboxylation, glucuronide conjugation
+
−
M91
8.17
C20H21O5
341.14029
341.14039
5.69
MS2[3 4 1]:281(1 0 0),237(20),219(10),209(6),297(5)
Dimethoxylation, hydroxylation
+
−
M92
8.17
C18H13O3
277.08582
277.08783
6.89
MS2[2 7 7]:233(1 0 0),259(20),249(11),219(5)
Dehydrogenation, hydroxylation
+
−
M93
8.17
C18H17O7S
377.06892
377.06985
2.39
MS2[3 7 7]:297(1 0 0),315(18),271(11),359(8)
Dihydroxylation, sulfonation
+
−
M94
8.21
C18H19O4
299.12771
299.13028
8.34
MS2[2 9 9]:239(1 0 0),281(3),221(3),133(1),263(1)
MS3[2 3 9]:221(1 0 0),219(19),133(11),198(1)Loss of C2H2, dimethoxylation
+
+
M95
8.21
C16H15O2
239.10667
239.10878
8.88
MS2[2 3 9]:221(1 0 0),133(27),219(10),211(3),237(2)
Loss of C2H2
+
+
M96
8.28
C16H13O2
237.09111
237.09271
7.19
MS2[2 3 7]:209(1 0 0),66(12),196(11),222(4),207(3)
MS3[2 0 9]:167(1 0 0),207(74),181(72),168(35)Loss of C2H2, dehydrogenation
+
−
M97
8.28
C18H17O3
281.11721
281.11908
6.61
MS2[2 8 1]:263(1 0 0),245(13),133(4),237(1)
MS3[2 6 3]:245(1 0 0),133(18),243(3),235(2)Hydroxylation
+
−
M98
8.28
C24H25O9
457.14931
457.15167
5.17
MS2[4 5 7]:281(1 0 0),439(68),295(55),175(6)
MS3[2 8 1]:263(1 0 0),245(15),133(9)Hydroxylation, glucuronide conjugation
+
−
M99
8.28
C19H21O5
329.13829
329.13983
4.50
MS2[3 2 9]:314(1 0 0),254(9),269(2),299(1)
MS3[3 1 4]:299(1 0 0),285(3),254(2)Loss of C2H2, trimethoxylation
+
−
M100
8.40
C14H9O3
225.05481
225.05637
7.77
MS2[2 2 5]:195(1 0 0),66(39),197(24),207(23),181(10)
Loss of C3H4 + CH2, dehydrogenation,
Hydroxylation+
−
M101
8.40
C18H17O6S
361.07401
361.07660
7.10
MS2[3 6 1]:281(1 0 0),305(8),343(7),331(1)
MS3[2 8 1]:263(1 0 0),245(13),133(2)Hydroxylation, sulfonation
+
−
M102
8.40
C20H21O5
341.14029
341.14120
8.35
MS2[3 4 1]:281(1 0 0),219(15),237(14),253(12)
Dimethoxylation, hydroxylation
+
−
M103
8.40
C15H11O5
271.06029
271.06219
7.71
MS2[2 7 1]:227(1 0 0),253(31),229(17),243(8)
Carboxylation, loss of C3H4, hydroxylation
+
−
M104
8.40
C16H13O6
301.07079
301.07254
6.23
MS2[3 0 1]:283(1 0 0),274(34),251(17),255(12)
Loss of 2CH2, tetrahydroxylation
+
−
M105
8.51
C18H13O3
277.08582
277.08838
8.88
MS2[2 7 7]:233(1 0 0),187(57),223(21),259(9)
Dehydrogenation, hydroxylation
+
−
M106
8.51
C24H25O10
473.14422
473.14780
7.56
MS2[4 7 3]:175(1 0 0),297(62),383(23),295(11)
Dihydroxylation, glucuronide conjugation
+
−
M107
8.61
C18H15O3
279.10151
279.10410
9.06
MS2[2 7 9]:261(1 0 0),233(26),231(21),259(8),205(4)
MS3[2 6 1]:233(1 0 0),231(57),207(28),205(14)Dehydrogenation, hydroxylation
+
−
M108
8.61
C19H21O5
329.13829
329.14050
6.53
MS2[3 2 9]:311(1 0 0),279(18),224(7),314(4)
MS3[3 1 1]:251(1 0 0),225(77),253(18)Loss of C2H2, trimethoxylation
+
−
M109
8.61
C18H13O2
261.09107
261.09326
8.63
MS2[2 6 1]:66(1 0 0),233(46),231(39),209(26)
Oxidation, dehydrogenation
+
−
M110
8.61
C19H19O5
327.12274
327.12515
7.49
MS2[3 2 7]:297(1 0 0),282(27),309(2)
MS3[2 9 7]:282(1 0 0),225(11),279(4),237(1)Loss of 2CH2, trimethoxylation
+
−
M111
8.61
C18H17O4S
329.08422
329.08221
−6.07
MS2[3 2 9]:315(1 0 0),249(39),301(18),302(16)
MS3[3 1 5]:235(1 0 0),274(77),288(18),208(3)Dehydroxylation, sulfonation
+
−
M112
8.72
C18H17O5
313.10701
313.10944
7.63
MS2[3 1 3]:283(1 0 0),251(60),295(54),267(39),285(30)
Loss of 2CH2, dimethoxylation, hydroxylation
+
−
M113
8.84
C18H17O4
297.11211
297.11490
9.30
MS2[2 9 7]:253(1 0 0),282(17),251(7),237(2),279(1)
Loss of 2CH2, dimethoxylation
+
−
M114
8.84
C17H17O2
253.12232
253.12457
8.94
MS2[2 5 3]:225(1 0 0),224(42),235(4)
MS3[2 2 5]:207(1 0 0),196(28),209(17)Loss of CH2, hydrogenation
+
−
M115
8.84
C19H19O5
327.12274
327.12540
8.25
MS2[3 2 7]:297(1 0 0),282(26),171(3),229(2)
MS3[2 9 7]:282(1 0 0),225(8)Loss of 2CH2, trimethoxylation
+
−
M116
8.96
C16H13O2
237.09111
237.09314
9.00
MS2[2 3 7]:209(1 0 0),196(35),207(26),222(11),219(3)
MS3[2 0 9]:193(1 0 0),168(83),181(44)Loss of C2H2, dehydrogenation
+
−
M117
8.96
C17H13O3
265.08581
265.08829
8.94
MS2[2 6 5]:237(1 0 0),207(14),247(12),196(5),209(4)
MS3[2 3 7]:209(1 0 0),207(58),235(34),193(18),196(12)Loss of CH2, dehydrogenation, hydroxylation
+
−
M118
8.96
C17H15O4
283.09641
283.09903
8.99
MS2[2 8 3]:239(1 0 0),265(61),221(51),237(31),225(14)
MS3[2 3 9]:221(1 0 0),133(67),219(21),237(11)Loss of CH2, dihydroxylation
+
−
M119
9.08
C17H15O3
267.10151
267.10379
8.31
MS2[2 6 7]:249(1 0 0),247(5),207(3),225(1),235(1)
MS3[2 4 9]:207(1 0 0),247(20),191(16),221(13)Loss of CH2, hydroxylation
+
−
M120
9.08
C16H15O3
255.10159
255.10365
8.15
MS2[2 5 5]:237(1 0 0),209(5),196(4),187(3))
MS3[2 3 7]:209(1 0 0),196(32),222(18),193(12)Loss of C2H2, hydroxylation
+
−
M121
9.08
C23H23O8
427.13877
427.14175
7.04
MS2[4 2 7]:295(1 0 0),277(8),391(2)
MS3[2 9 5]:251(1 0 0),233(6),277(1)Carboxylation, glucuronide conjugation,
decarboxylation+
−
M122
9.30
C18H17O4
297.11211
297.11429
7.25
MS2[2 9 7]:237(1 0 0),225(75),267(41),279(25),261(11)
MS3[2 3 7]:209(1 0 0),223(18),219(8)Loss of 2CH2, dimethoxylation
+
−
M123
9.42
C16H13O4
269.08092
269.08319
8.75
MS2[2 6 9]:225(1 0 0),207(10),251(4),181(3)
MS3[2 2 5]:207(1 0 0),205(30)Loss of 2CH2, dihydroxylation
+
−
M124
9.53
C17H15O3
267.10151
267.10400
9.09
MS2[2 6 7]:249(1 0 0),239(36),133(15),221(13),247(4)
MS3[2 4 9]:249(1 0 0),221(96),247(47)Loss of CH2, hydroxylation
+
−
M125
9.65
C15H13O4
257.08099
257.08307
8.69
MS2[2 5 7]:239(1 0 0),109(71),163(39),242(30),213(28),224(9)
MS3[2 3 9]:145(1 0 0),224(77),221(48),211(47),196(30)Loss of C2H2 + CH2, dihydroxylation
+
−
M126
9.65
C16H15O5
287.09149
287.09363
7.77
MS2[2 8 7]:272(1 0 0),269(5),219(2)
MS3[2 7 2]:152(1 0 0),123(8),257(5)Loss of C3H4 + CH2, dimethoxylation,
hydroxylation+
−
M127
9.76
C18H15O2
263.10661
263.10895
8.72
MS2[2 6 3]:245(1 0 0),133(13),235(2),221(1)
MS3[2 4 5]:245(1 0 0),243(48),218(37)Dehydrogenation
+
−
M128
9.87
C18H15O3
279.10151
279.10416
9.28
MS2[2 7 9]:261(1 0 0),233(25),231(21),259(7),205(4)
MS3[2 6 1]:233(1 0 0),231(68),217(37),205(10)Dehydrogenation, hydroxylation
+
−
M129
9.87
C16H15O5
287.09149
287.09366
7.87
MS2[2 8 7]:269(1 0 0),272(61),219(28),258(13),227(3)
Loss of C3H4 + CH2, dimethoxylation,
hydroxylation+
−
M130
9.87
C23H24O6NS
442.13192
442.13541
7.97
MS2[4 4 2]:279(1 0 0),162(2),313(1)
MS3[2 7 9]:261(1 0 0),233(27),231(23),259(10)Hydroxylation, N-acetylcysteine conjugation
+
−
M131
9.98
C17H15O3
267.10151
267.10312
5.80
MS2[2 6 7]:249(1 0 0),239(23),133(9),221(7),247(2)
Loss of CH2, hydroxylation
+
−
M132
9.98
C17H17O3
269.11729
269.11856
4.98
MS2[2 6 9]:239(1 0 0),207(26),251(17),221(15),225(8)
Loss of C2H2, methoxylation
+
−
M133
10.09
C17H15O2
251.10661
251.10831
6.59
MS2[2 5 1]:233(1 0 0),231(33),221(5),247(1)
MS3[2 3 3]:191(1 0 0),187(69),207(52),203(29)Loss of CH2
+
−
M134
10.20
C19H19O5S
359.09482
359.09735
7.18
MS2[3 5 9]:279(1 0 0),344(39),264(31),261(12)
Methylation, sulfonation
+
−
M135
10.64
C17H15O2
251.10661
251.10841
6.98
MS2[2 5 1]:233(1 0 0),231(9),210(1),223(1)
MS3[2 3 3]:231(1 0 0),207(19),215(8),192(5)Loss of CH2
+
−
M136
10.64
C18H17O3
281.11721
281.11923
7.15
MS2[2 8 1]:263(1 0 0),245(45),133(7),233(5),237(2)
MS3[2 6 3]:245(1 0 0),133(8),243(6),235(3)Hydroxylation
+
+
M137
10.64
C18H15O4
295.09641
295.09866
7.37
MS2[2 9 5]:251(1 0 0),233(7),231(1),267(1)
MS3[2 5 1]:233(1 0 0),231(9),210(1)Carboxylation
+
−
M138
10.64
C18H17O4
297.11211
297.11380
5.60
MS2[2 9 7]:297(1 0 0),279(47),224(14),282(11),267(1)
Loss of 2CH2, dimethoxylation
+
−
M139
10.64
C18H13O3
277.08582
277.08774
6.57
MS2[2 7 7]:233(1 0 0),205(60),259(32),217(17)
Dehydrogenation, hydroxylation
+
−
M140
10.89
C16H13O2
237.09111
237.09305
8.62
MS2[2 3 7]:209(1 0 0),196(11),66(7),164(5),207(4)
MS3[2 0 9]:207(1 0 0),193(49),181(37)Loss of 2CH2
+
−
M141
10.89
C18H17O4
297.11211
297.11462
8.36
MS2[2 9 7]:225(1 0 0),237(90),279(38),253(35),235(27)
MS3[2 2 5]:207(1 0 0),205(14)Loss of 2CH2, dimethoxylation
+
−
M142
10.89
C20H21O5
341.14029
341.14050
6.30
MS2[3 4 1]:281(1 0 0),237(20),299(6),209(5),313(2)
MS3[2 8 1]:237(1 0 0),261(9),239(6),253(4)Dimethoxylation, hydroxylation
+
−
M143
10.89
C17H17O2
253.12232
253.12433
8.00
MS2[2 5 3]:235(1 0 0),207(23),233(7),212(2)
MS3[2 3 5]:207(1 0 0),233(14),205(10)Loss of CH2, hydrogenation
+
−
M144
11.00
C19H19O5
327.12274
327.12509
7.31
MS2[3 2 7]:312(1 0 0),309(33),268(24),291(15)
MS3[3 1 2]:297(1 0 0),282(59),271(7),241(4)Loss of 2CH2, trimethoxylation
+
−
M145
11.02
C18H15O2
263.10661
263.10904
9.06
MS2[2 6 3]:245(1 0 0),133(23),235(5),243(3)
MS3[2 4 5]:245(1 0 0),217(71),243(53),204(50)Dehydrogenation
−
+
M146
11.02
C18H19O3
283.13291
283.13510
7.87
MS2[2 8 3]:265(1 0 0),217(24),263(17),247(15),255(14)
MS3[2 6 5]:247(1 0 0),245(44),220(15)Hydration
+
+
M147
11.02
C17H15O
235.11177
235.11391
9.22
MS2[2 3 5]:194(1 0 0),66(74),157(69),220(33),200(22)
Loss of CH2, dehydroxylation
−
+
M148
11.11
C18H17O3
281.11721
281.11951
8.14
MS2[2 8 1]:263(1 0 0),245(23),133(8),235(1)
MS3[2 6 3]:245(1 0 0),133(29),235(5),243(2)Hydroxylation
+
+
M149
11.22
C19H19O4
311.12772
311.13019
7.73
MS2[3 1 1]:293(1 0 0),211(21),275(19),183(17),296(13)
MS3[2 9 3]:278(1 0 0),275(98),249(51),171(14)Methoxylation, hydroxylation
+
−
M150
11.24
C18H15O2
263.10661
263.10901
8.95
MS2[2 6 3]:245(1 0 0),133(8),243(5),235(2)
MS3[2 4 5]:218(1 0 0),245(92),243(85),217(53)Dehydrogenation
+
+
M151
11.34
C16H13O3
253.08601
253.08818
8.93
MS2[2 5 3]:235(1 0 0),207(23),233(7),212(2),225(1)
MS3[2 3 5]:207(1 0 0),233(14),208(13),194(5)Loss of 2CH2, hydroxylation
+
−
M152
11.34
C18H15O7
343.08342
343.08380
7.49
MS2[3 4 3]:325(1 0 0),299(55),315(14),271(9)
Carboxylation, trihydroxylation
+
−
M153
11.57
C17H15O3
267.10151
267.10403
9.21
MS2[2 6 7]:249(1 0 0),247(6),207(3),226(1)
MS3[2 4 9]:207(1 0 0),206(47),221(3)Loss of CH2, hydroxylation
+
−
M154
12.02
C18H17O3
281.11721
281.11981
9.21
MS2[2 8 1]:263(1 0 0),245(38),133(6),233(4),237(3)
MS3[2 6 3]:245(1 0 0),133(16),243(4),219(2)Hydroxylation
+
−
M155
12.08
C24H25O8
441.15441
441.15805
8.29
MS2[4 4 1]:265(1 0 0),175(10),307(2),176(1)
MS3[2 6 5]:247(1 0 0),245(24),243(1),224(1)Glucuronide conjugation
+
+
M156
12.08
C18H17O2
265.12229
265.12479
9.37
MS2[2 6 5]:247(1 0 0),265(24),245(19),224(1)
MS3[2 4 7]:245(1 0 0),247(61),204(57),221(54)Magnolol isomer
+
+
M157
12.13
C17H15O2
251.10661
251.10884
8.70
MS2[2 5 1]:233(1 0 0),231(10),189(2),133(1)
Loss of CH2
+
−
M158
12.13
C15H11O4
255.06532
255.06746
8.92
MS2[2 5 5]:213(1 0 0),211(42),187(19),227(7),193(4)
Loss of C3H4, dehydrogenation,
dihydroxylation+
−
M159
12.31
C18H15O3
279.10151
279.10406
8.92
MS2[2 7 9]:261(1 0 0),233(82),259(4),251(3)
MS3[2 6 1]:233(1 0 0),259(6),219(5),218(4)Dehydrogenation, hydroxylation
−
+
M160
12.36
C18H13O2
261.09107
261.09320
8.40
MS2[2 6 1]:233(1 0 0),66(8),179(2)
MS3[2 3 3]:231(1 0 0),207(51),205(48)Oxidation, dehydrogenation
+
−
M161
12.47
C16H11O5
283.06019
283.06259
8.80
MS2[2 8 3]:268(1 0 0),213(4),250(1)
MS3[2 6 8]:267(1 0 0),239(42),240(36),224(31)Loss of C2H2, oxidation, carboxylation,
hydroxylation+
−
M162
12.63
C18H17O5S
345.07911
345.08191
8.08
MS2[3 4 5]:265(1 0 0),301(5),283(2),246(1)
MS3[2 6 5]:247(1 0 0),245(30),224(3)Sulfonation
−
+
M163
12.65
C18H17O2
265.12229
265.12482
9.48
MS2[2 6 5]:247(1 0 0),245(19),217(2),224(1)
MS3[2 4 7]:245(1 0 0),219(59)Magnolol isomer
−
+
M164
12.91
C15H11O5
271.06029
271.06247
8.74
MS2[2 7 1]:256(1 0 0),66(22),225(13),209(6)
Loss of C2H2 + CH2, dehydrogenation, trihydroxylation
+
−
M165
13.59
C18H17O3
281.11721
281.11996
9.74
MS2[2 8 1]:263(1 0 0),261(20),207(4),252(1)
MS3[2 6 3]:206(1 0 0),207(90),233(86),231(69)Hydroxylation
+
−
M166
13.60
C28H32O11N3S2
650.14732
650.14221
−7.79
MS2[6 5 0]:570(1 0 0),305(45),345(29),265(7)
Sulfonation, glutathione conjugation
−
+
M167
14.39
C19H19O4
311.12772
311.13040
8.40
MS2[3 1 1]:293(1 0 0),237(15),209(12),291(5)
MS3[2 9 3]:249(1 0 0),275(38),247(16)Methoxylation, hydroxylation
+
−
M168
14.50
C19H17O4
309.11219
309.11499
9.23
MS2[3 0 9]:291(1 0 0),233(39),261(25),277(17),265(10)
MS3[2 9 1]:263(1 0 0),231(80),273(44),233(30),247(20)Carboxylation, methylation
+
−
M169
14.62
C18H17O6
329.10189
329.10278
2.48
MS2[3 2 9]:311(1 0 0),201(32),293(18),217(7),281(6)
Tetrahydroxylation
+
−
M170
14.87
C19H17O4
309.11219
309.11484
8.75
MS2[3 0 9]:291(1 0 0),247(23),193(11),265(7),209(6)
MS3[2 9 1]:247(1 0 0),273(93),193(37),229(18),275(6)Carboxylation, methylation
+
−
M171
17.34
C15H11O2
223.07551
223.07654
5.31
MS2[2 2 3]:205(1 0 0),195(9),179(3),207(2)
Loss of C3H4, dehydrogenation
−
+
M172
17.34
C18H15O
247.11181
247.11307
5.38
MS2[2 4 7]:206(1 0 0),220(14),245(8)
Dehydration
+
+
M173
17.34
C28H30O9N3S
584.16969
584.16510
−7.92
MS2[5 8 4]:279(1 0 0),305(76),582(48),265(6)
Carboxylation, glutathione conjugation
+
+
M174
17.34
C17H15O
235.11177
235.11293
5.05
MS2[2 3 5]:66(1 0 0),200(28),157(25),202(21)
Loss of CH2, dehydroxylation
−
+
M175
17.34
C29H32O10NS
586.17419
586.17169
−4.19
MS2[5 8 6]:526(1 0 0),371(10),441(5),175(1)
Glucuronide conjugation, N-acetylcysteine conjugation
−
+
M176
27.75
C22H19O10
443.09739
443.09583
−3.26
MS2[4 4 3]:267(1 0 0),175(33),249(21),223(13)
Carboxylation, Loss of C2H2, dehydrogenation, glucuronide conjugation
−
+
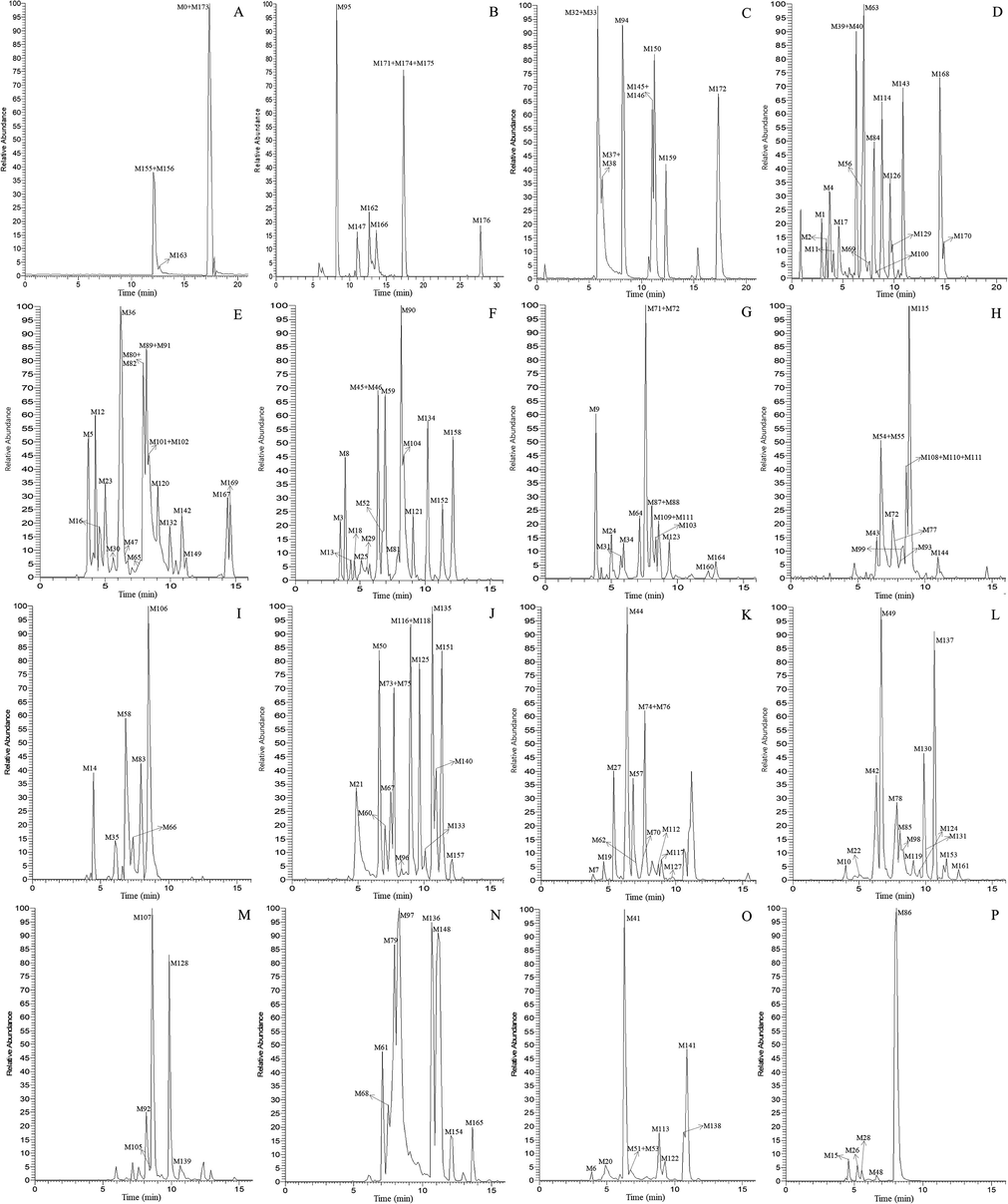
High resolution extracted ion chromatograms for the multiple daidzein metabolites in rat urine and plasma (A-C for plasma and D-P for urine): (A) m/z 223.07551, 235.11177, 239.10667, 345.07911, 443.09739, 586.17419, 650.14732; (B) m/z 247.11181, 263.10661, 279.10151, 283.13291, 299.12771, 379.08459; (C) m/z 265.12229, 441.15441, 584.16969; (D) m/z 209.05991, 225.05481, 243.06539, 253.12232, 287.09149, 309.11219, 313.07059, 333.13319, 395.07949; (E) m/z 255.10159, 269.11729, 311.12772, 315.12269, 329.10189, 341.14029, 361.07401; (F) m/z 255.06532, 301.07079, 343.08342, 359.09482, 415.10249, 427.13877, 461.18059, 471.12859; (G) m/z 261.09107, 269.08092, 271.06029, 329.08422, 400.12132, 429.08179, 637.17634; (H) m/z 327.12274, 329.08422, 329.13829, 377.06892; (I) m/z 473.14422; (J) m/z 237.09111, 251.10661, 253.08601, 257.08099, 283.09641, 333.00652; (K) m/z 263.10661, 265.08581, 313.10701, 475.15989; (L) m/z 267.10151, 283.06019, 295.09641, 442.13192, 457.14931; (M) m/z 277.08582, 279.10151; (N) m/z 281.11721; (O) m/z 253.04971, 297.11211; (P) m/z 269.04459.
3.4.1 Identification of metabolites M0, M156 and M163
In negative ion mode, the metabolites M0, M156 and M163 exhibited the same theoretical [M−H]- ions at m/z 265.12229 (C18H17O2, error ≤ 9.50 ppm), which were eluted at 17.34, 12.08 and 12.65 min, respectively. Based on the comparison of the chromatographic retention time and MS/MSn spectral data with the magnolol reference standard, M0 could be accurately identified as magnolol (Guo et al., 2019; Wu et al., 2006; Zhu et al., 2020). Since the major product ions of M156 and M163 (m/z 247, 245 and 224) were consistent with M0, they could be inferred as positional isomers of magnolol.
3.4.2 Identification of metabolites M32, M37, M47, M65, M82, M94, M95, M120, M132 and their secondary metabolites
M95, eluted at 8.21 min, was 26 Da lower than magnolol, which was presumed to be the product of magnolol lost CH = CH. In its ESI-MS2 spectrum, a series of NLFs similar to magnolol were observed, such as 2 Da (m/z 239 → m/z 237), 18 Da (m/z 239 → m/z 221) and 28 Da (m/z 239 → m/z 211), which further confirmed our inference. Metabolites M47 and M120 had the same [M−H]- ions at m/z 255.10159 (C16H15O3, error ≤ 9.50 ppm), which was 16 Da higher than M95. Therefore, they were considered to be hydroxylation products of M95 because of their similar fragmentation behaviors. In the same way, M65 M82 and M132 (C17H17O3, 30 Da higher than M95) were identified as methoxylated products of M95, while M32, M37 and M94 (C18H19O4, 60 Da higher than M95) were inferred as di-methoxylated products of M95.
Meanwhile, in the HREIC, M43, M54, M99 and M108 were extracted at m/z 329.13829 (C19H21O5, error ≤ 7.00 ppm) with retention times ranging from 6.40 to 8.61 min. They were 90 Da higher than M95, and the continuous NLFs of 30 Da (m/z 329 → m/z 299 → m/z 269 → m/z 239) occurred in the ESI-MS2 spectra, which further confirmed that they were tri-methoxylated products of M95. Metabolites M5, M12, M16, M30 and M36 produced the same [M−H]- ions at m/z 315.12269 (C18H19O5, error ≤ 9.50 ppm), which were 16 Da higher than that of M32. Furthermore, the identical ion fragments with NLFs indicated that they were hydroxylation products of M32. In the same vein, M33 and M38 (m/z 379.08459, C18H19O7S), M25, M29 and M59 (m/z 461.18059, C24H29O9), M27, M44 and M57 (m/z 475.15989, C24H27O10) were 80 Da, 162 Da and 176 Da higher than M32, respectively. In their ESI-MS2 spectra, the same DPIs at m/z 299 ([M−SO3−H]-, [M−C6H10O5−H]- and [M−C6H8O6−H]-) demonstrated that they were sulfonated, glucose conjugated, and glucuronide conjugated products of M32, respectively.
The metabolite M24, with a retention time of 5.00 min, yielded a [M−H]- ion at m/z 637.17634 (C29H33O16, 7.41 ppm), which was 162 Da higher than that of M27. In the ESI-MSn spectrum, the fragment ions appeared including [M−GluA−H]- at m/z 461 and [M−GluA−Glc−H]- at m/z 299, both indicated that M24 was the glucose conjugated product of M27. M4 and M17 were eluted at 3.67 min and 4.57 min separately, and possessed the same [M−H]- ion at m/z 395.07949 (C18H19O8S, error ≤ 8.50 ppm), which were 80 Da higher than M5. Therefore, M4 and M17 were inferred as the sulfonated products of M5.
Metabolites M50 and M125 were eluted at 6.62 min and 9.65 min, respectively. Both of them gave rise to a deprotonated molecular ion with the molecular formula C15H13O4 at m/z 257.08099. Their ESI-MS2 base peak ions at m/z 239 and m/z 242 were individually formed due to neutral loss of H2O and CH3, suggesting that they might be products of di-hydroxylated magnolol lost C2H2 + CH2.
3.4.3 Identification of metabolites M61, M68, M79, M97, M136, M148, M154, M155, M162, M165 and their secondary metabolites
M61, M68, M79, M97, M136, M148, M154, and M165 were detected at 7.05 min, 7.49 min, 7.95 min, 8.28 min, 10.64 min, 11.11 min, 12.02 min, and 13.59 min, respectively, and had the same [M−H]- ion located at m/z 281.11721 (C18H17O3, error ≤ 10.00 ppm). All of them were 16 Da higher than magnolol, indicating that they could be identified as hydroxylation products of magnolol. In their ESI-MSn spectra, the emergence of multiple identical NLFs, such as 18 Da (m/z 281 → m/z 263), 2 Da (m/z 263 → m/z 261) and 28 Da (m/z 263 → m/z 235), further corroborated the above conclusion.
M155 was eluted at 12.08 min and produced its [M−H]- ion at m/z 441.15441 (C24H25O8, 8.29 ppm). It was 176 Da higher than magnolol, and [M−GluA−H]- at m/z 265 was also observed, so M155 was presumed to be the glucuronidation product of magnolol. Along the same lines and evidence, M162 could be identified as the sulfonated product of magnolol.
The metabolites M31 and M64 were eluted at 5.73 min and 7.17 min, and generated the same deprotonated molecular ion at m/z 400.12132 (C21H22O5NS, error ≤ 8.50 ppm). In their ESI-MS2 spectra, the [M−C3H6NO2S−H]- at m/z 279 were both detected, indicating that they might be the N-acetylcysteine conjugation products of M61. In the same manner, M78, M85 and M98 (m/z 457.14931, C24H25O9), M80, M89 and M101 (m/z 361.07401, C18H17O6S), M130 (m/z 442.13192, C23H24O6NS), M149 and M167 (m/z 311.12772, C19H19O4), M23 and M169 (m/z 329.10189, C18H17O6) could be identified as the glucuronidation, sulfonation, N-acetylcysteine conjugation, methoxylation, and tri-hydroxylation products of M61, respectively.
All metabolites M14, M35, M66, M83 and M106 yielded the same [M−H]- ions at m/z 473.14422 (C24H25O10, error ≤ 8.00 ppm) and were 16 Da higher than M78, which proved that they were hydroxylated products of M78. In their ESI-MS2 spectra, there were abundant product ions at m/z 297 due to the neutral loss of 176 Da, which provided an adequate basis for our deduction. Similarly, M77 and M93 (m/z 377.06892, C18H17O7S) could be presumed as the hydroxylated products of M80, while M91, M102 and M142 (m/z 341.14029, C20H21O5) were characterized as the methoxylated products of M149.
In negative ion mode, M53 was eluted at 6.73 min and its elemental composition was proposed to be C18H21O4 (6.59 ppm). In its MS/MS spectrum, a series of characteristic product ions were observed, such as m/z 283 ([M−H2O−H]-), m/z 240 ([M−H2O−C3H7−H]-), m/z 273 ([M−CO−H]-), and m/z 258 ([M−CO−CH3−H]-), so that M53 was clearly identified as the hydrogenation and dihydroxylation product of magnolol.
3.4.4 Identification of metabolites M42, M46, M75, M118, M119, M124, M131, M153 and their secondary metabolites
The metabolites M42, M119, M124, M131 and M153 were 14 Da lower than that of M61 with retention times of 6.29 min, 9.08 min, 9.53 min, 9.98 min and 11.57 min, suggesting that they might be the products of M61 lost CH2. The NLFs of 18 Da, 28 Da and 2 Da were consistent with the MS/MS spectrum of magnolol, which validated our above hypothesis. M75 and M118 owned the same [M−H]- ion at m/z 283.09641 (C17H15O4, error ≤ 9.50 ppm), and they were 16 Da higher than M42, which meant that they were hydroxylation products of M42. Similarly, M46 could be identified as the di-methoxylated product of M75.
M140 was eluted at 10.89 min and gave rise to its [M−H]- ion at m/z 237.09111 (C16H13O2, 8.62 ppm), which was 28 Da lower than magnolol. Based on the predicted molecular formula, it could be inferred that M140 was the product of magnolol lost 2CH2. The metabolite M151 was 16 Da higher than M140, which illustrated that it was the hydroxylation product of M140. According to the established line of reasoning, M88 and M123 were the di-hydroxylated products of M140, while M51, M113, M122, M138 and M141 were the di-methoxylated products of M140 (as speculated by the continuous NLFs of 30 Da).
M55, which was eluted at 6.73 min, was 80 Da higher than M51 in negative ion mode, suggesting that it might be the sulfonation product of M51. In the MS2 spectra, the neutral loss of 80 Da (m/z 377 → m/z 297) and the fragmentation behaviors of m/z 297 both provided ample support for our inference. With this method for continuous practice, M58 (176 Da higher than M51), M110, M115 and M144 (30 Da higher than M51), M7, M19, M62, M76 and M112 (16 Da higher than M51) could be individually identified as glucuronidation, methoxylation and hydroxylation products of M51, while M104 (32 Da higher than M88) was reckoned as dihydroxylation product of M88.
The metabolite M69 exhibited its [M−H]- ion at m/z 243.06539 (C14H11O4, 8.29 ppm), which was 26 Da lower than M88. In the ESI-MS2 spectrum, the fragmentation behaviors of M69 were in line with that of M88, so it could be deduced that M69 was the product of M88 lost CH = CH. Metabolites M11, M56, M126 and M129, which were separately eluted at 4.08 min, 6.84 min, 9.65 min, and 9.87 min, possessed the same theoretical [M−H]- ions at m/z 287.09149 (C16H15O5, error ≤ 9.50 ppm). In their MS/MS spectra, the DPI at m/z 227 and NLFs of 15 Da (m/z 287 → m/z 272 → m/z 257) were both detected, implying that two molecules of methoxy were introduced into the structure. For this reason, M11, M56, M126 and M129 were identified as the de-hydroxylation and dimethoxylation products of M69.
M39 was eluted at 6.29 min and yielded a [M−H]- ion at m/z 209.05991 (C14H9O2, 7.10 ppm). It was 28 Da lower than M140, which revealed that M39 was the dehydrogenation product of M140 lost CH = CH. Metabolites M40, M84, and M100 with the same [M−H]- ion at m/z 225.05481 (C14H9O3, error ≤ 8.00 ppm) were eluted at 6.29 min, 8.06 min, and 8.40 min, respectively. All of them were 16 Da higher than M39, so these three isomers could be characterized as the hydroxylation products of M39. At the same time, M18 and M45 (m/z 255.06532, C15H11O4) were identified as the hydroxylation and methoxylation products of M39.
M60, M67, M133, M135 and M157 were individually eluted at 7.05 min, 7.49 min, 10.09 min, 10.64 min and 12.13 min with the same [M−H]- ion at m/z 251.10661 (C17H15O2, error ≤ 10.00 ppm), which were 14 Da lower than magnolol and its isomers (M156, M163). Hence, it could be presumed that they were the products of M0, M156 and M163 lost CH2, and they were isomers because the substitution sites of two hydroxyl were different. In the same way, M147 and M174 (C17H15O, 16 Da lower than M60) were inferred as the dehydroxylated products of M60.
The retention times of M74 and M117 were 7.72 min and 8.96 min, respectively, which were 2 Da lower than that of M42, suggesting that they were dehydrogenation products of M42. Metabolites M114 and M143 were separately eluted at 8.84 min and 10.89 min, and produced the same deprotonated molecular ion at m/z 253.12232 (C17H17O2, error ≤ 9.00 ppm), which were 12 Da lower than that of magnolol. By comparing with the MS/MS spectral data of magnolol, M114 and M143 could be identified as the hydrogenation products of magnolol lost CH2. In the RP-HPLC system, the compounds with larger Clog P values will be more difficult to be eluted. Therefore, M114 (Clog P, 4.20) and M143 (Clog P, 4.38) were isomers to each other.
3.4.5 Identification of metabolites M52, M81, M90, M137, M152, M168, M170 and their secondary metabolites
The metabolite M137 eluted at 10.64 min generated a [M−H]- ion at m/z 295.09641 (C18H15O4, 7.37 ppm), which was 30 Da higher than that of magnolol. In its ESI-MS2 spectrum, M137 exhibited a NLF of 44 Da, resulting in the production of [M−CO2−H]- ion at m/z 251, which was attributed to the presence of carboxyl group in its structure. By comparing the mass fragmentation behaviors of m/z 251 in the MS3 spectra with that of magnolol, M137 could be identified as the carboxylation product of magnolol. The metabolites M52, M81 and M90 were separately eluted at 6.73 min, 7.95 min and 8.17 min, and gave rise to the same [M−H]- ions at m/z 471.12859 (C24H23O10, error ≤ 6.50 ppm). Their ESI-MSn spectra all showed the DPIs at m/z 295 ([M−GluA−H]-), which could help us to infer that they were glucuronidation products of M137. With the same inference thinking, M152 (C18H15O7, 48 Da higher than M137) were identified as the tri-hydroxylated product of M137, while M168 and M170 (C19H17O4, 14 Da higher than M137) could be presumed to be methylation products of M137.
The metabolite M121 eluted at 9.08 min exhibited its [M−H]- ion at m/z 427.13877 (C23H23O8, 7.04 ppm), which was 132 Da higher than M137 or 44 Da lower than M52. In the MS2 spectra, M121 produced a DPI at m/z 295 through the neutral loss of 132 Da (C5H8O4), which proved that M121 was the decarboxylation product of M52. M173 displayed a [M−H]-ion at m/z 584.16969 (C28H30O9N3S, −7.92 ppm), which was 289 Da higher than that of M137. The NLF of 305 Da (m/z 584 → m/z 279) suggested that M173 might be the glutathione conjugation product of M137 and that the binding site was at the carboxyl group.
Metabolites M71, M87, and M103, which possessed the same [M−H]- ions at m/z 271.06029 (C15H11O5, error ≤ ±9.00 ppm), were eluted at 7.60 min, 8.06 min, and 8.40 min, respectively. By comparing with the molecular formula and atomic occupancy of M137 and combining with the MSn spectral data, M71, M87, and M103 could be surmised as de-propenyl (C3H4) and hydroxylation products of M137. The metabolite M176 eluted at 27.75 min showed a [M−H]- ion at m/z 443.09739 (C22H19O10, −3.26 ppm), and then produced the DPIs at m/z 267 and m/z 223 through the continuous neutral loss of 176 Da (GluA) and 44 Da (CO2), suggesting that M176 were de-vinyl (C2H2) and dehydrogenation product of M52.
Metabolites M10, M22, M49, and M161 were extracted from HREICs at m/z 283.06019 (C16H11O5, error ≤ 10.00 ppm) in negative ion mode with retention times of 3.97 min, 5.00 min, 6.62 min, and 12.47 min. None of them had fragment ions at m/z 265, so the hydroxyl group in the molecules might be oxidized. In their MSn spectra, the NLFs of 15 Da (m/z 283 → m/z 268), 28 Da (m/z 283 → m/z 255) and 44 Da (m/z 268 → m/z 224) were all observed, which demonstrated that M10, M22, M49 and M161 were oxidation, de-vinyl (C2H2) and hydroxylation products of M137. Based on this assertion, M63 (C17H13O6, 30 Da higher than M10) was identified as the methoxylation product of M10, while M15, M26, M28, M48 and M86 (C15H9O5, 14 Da lower than M10) could be reckoned as the demethylation products of M10.
3.4.6 Identification of metabolites M107, M128, M159 and their secondary metabolites
Metabolites M107, M128, and M159 were eluted at 8.61 min, 9.87 min, and 12.31 min, respectively. All of them yielded the same [M−H]- ions at m/z 279.10151 (C18H15O3, error ≤ 9.50 ppm), which were 14 Da higher than M0 or 2 Da lower than M61. In their ESI-MS2 spectra, the DPI at m/z 261 was also 2 Da lower than the DPI at m/z 263 generated by M61, suggesting that M107, M128, and M159 were dehydrogenation products of M61. In the same way, M92, M105 and M139 (C18H13O3, 277.08582) could be identified as the dehydrogenation products of M107.
The metabolites M73, M96 and M116, whose retention times were 7.72 min, 8.28 min and 8.96 min, produced the same [M−H]- ions at m/z 237.09111 (C16H13O2, error ≤ 9.50 ppm, 28 Da lower than M0). The [M−CH3−H]- ions at m/z 222 were unreservedly shown in their ESI-MS2 spectra, which indicated that M73, M96 and M116 were the de-vinyl (C2H2) and dehydrogenation products of M0. The metabolite M164 eluted at 12.91 min possessed a [M−H]- ion at m/z 271.06029 (C15H11O5, 8.74 ppm), which was 34 Da higher than that of M73. The NLF of 15 Da (m/z 271 → m/z 256) was observed in its MS2 spectrum, so M164 could be speculated as the de-methylene and tri-hydroxylation product of M73.
The metabolite M171 eluted at 17.34 min gave rise to a [M−H]- ion at m/z 223.07551 (C15H11O2, 5.31 ppm), which was 14 Da lower than M73. A DPI at m/z 205 was monitored, indicating that M171 was the de-propenyl (C3H4) and dehydrogenation product of magnolol. On the basis of the above inference, M158 (C15H11O4, 255.06532, 32 Da higher than M171) was identified as the di-hydroxylation product of M171.
Metabolites M6, M20, and M41 with retention times of 3.87 min, 4.88 min, and 6.29 min, respectively, showed identical [M−H]- ions at m/z 253.04971 (C15H9O4, error ≤ 10.00 ppm), which were 2 Da lower than M158. In their MS2 spectra, the NLFs of 28 Da (m/z 253 → m/z 225 → m/z 197) provided sufficient evidence for inferring that they were oxidation products of M158. Metabolites M3, M8, and M13 owned the same [M−H]- ions at m/z 415.10249 (C21H19O9, error ≤ 9.00 ppm). In the ESI-MSn spectra, there were abundant base peak ions at m/z 253 due to the neutral loss of 162 Da (C6H10O5), suggesting that they were glucose conjugation products of M6. In the same manner, M9 (C21H17O10, 176 Da higher than M6) was inferred as the glucuronidation product of M6, while M21 (C15H9O7S, 80 Da higher than M6) could be identified as the sulfonation product of M6.
3.4.7 Identification of secondary metabolites of metabolites M0, M156 and M163
Metabolite M172, 18 Da lower than M0, exhibited a deprotonated molecular ion at m/z 247.11181 (C18H15O, 5.38 ppm) with the retention time of 17.34 min. The DPIs at m/z 245, 220 and 206 pointed out that fragmentation behaviors of M172 were similar to that of magnolol, and thus M172 was identified as the dehydrated product of magnolol. Conversely, M146 (C18H19O3, 18 Da higher than M0) was inferred to be the hydrated product of magnolol.
Metabolites M1 and M2 were separately eluted at 2.92 and 3.39 min, and yielded the same [M−H]- ion at m/z 333.13319 (C18H21O6, error ≤ 8.00 ppm) in negative ion mode. The consecutive NLFs of 60 Da (m/z 333 → m/z 273 → m/z 213) implied that four methoxy groups were introduced into the molecules. Therefore, M1 and M2 were diagnosed as the tetra-methoxylation products of magnolol lost 2C2H2. M72 and M111(C18H17O4S, 329.08422), whose retention times were 7.60 min and 8.61 min, were 16 Da lower than that of M162, suggesting that both were de-hydroxylation and sulfonation products of M0, M156 and M163.
The metabolites M70, M127, M145 and M150 were individually detected at 7.60 min, 9.76 min, 11.02 min and 11.24 min with the same [M−H]- ions at m/z 263.10661 (C18H15O2, error ≤ 9.50 ppm), which were 2 Da lower than M0. By comparing with the mass fragmentation behaviors of M0, M156 and M163, they could be tentatively identified as the dehydrogenation products of magnolol and its isomers.
M34, M109 and M160 gave rise to a deprotonated ion at m/z 261.09107 (C18H13O2, error ≤ 9.00 ppm), which were eluted at 5.95 min, 8.61 min and 12.36 min, respectively. In the MSn spectra, the DPIs at m/z 233 and m/z 205 were observed owing to continuous neutral loss of CO (28 Da), which validated that the hydroxyl groups in the molecules were oxidized. Accordingly, M34, M109 and M160 were inferred as the oxidation and dehydrogenation products of M0, which were isomers of each other.
The metabolite M134 generated the [M−H]- ion at m/z 359.09482 (C19H19O5S, 7.18 ppm) with a retention time of 10.20 min. It was 14 Da higher than M162, and the DPIs at m/z 279 ([M−SO3−H]-) and m/z 264 ([M−SO3−CH3−H]-) in its ESI-MS2 spectrum further fully confirmed that M134 was the methylation product of M162. In the same way, M166 (C28H32O11N3S2, 305 Da higher than M162) was determined as the glutathione conjugation product of M162, while M175 (C29H32O10NS, 145 Da higher than M155) could be deduced as the N-acetylcysteine conjugation product of M155.
3.5 Proposed biotransformation pathways of magnolol in rats
In the present study, a total of 177 metabolites (prototype compound included) with different molecular structures were monitored and identified in rats after oral administration of magnolol. As a beneficial phenolic compound, the in vivo biotransformation process of magnolol could be summed up as undergoing three fundamental metabolic pathways. The first metabolic pathway (shown in Fig. 6): parent drug and its isomers were bio-transformed into drug metabolism centres(Liu et al., 2020; Mei et al., 2019), such as hydroxylated magnolol, de-methylene magnolol and de-vinyl magnolol, based on which many chemical reactions occurred to generate a series of secondary metabolites. The second metabolic pathway (shown in Fig. 7): magnolol was first metabolized into carboxylated magnolol and dehydrogenated magnolol, both of which served as metabolic centers to gradually produced final metabolites. The third metabolic pathway (shown in Fig. 8): magnolol and its isomers went through intricate biotransformation processes, such as methoxylation, sulfonation, oxidation, and hydration, to yield complicated secondary metabolites. It was noteworthy that while both phase I and phase II metabolic reactions were observed, the vast majority of in vivo metabolites were generated either through binding reactions (phase II) or composite reactions (including various types of biotransformation reactions).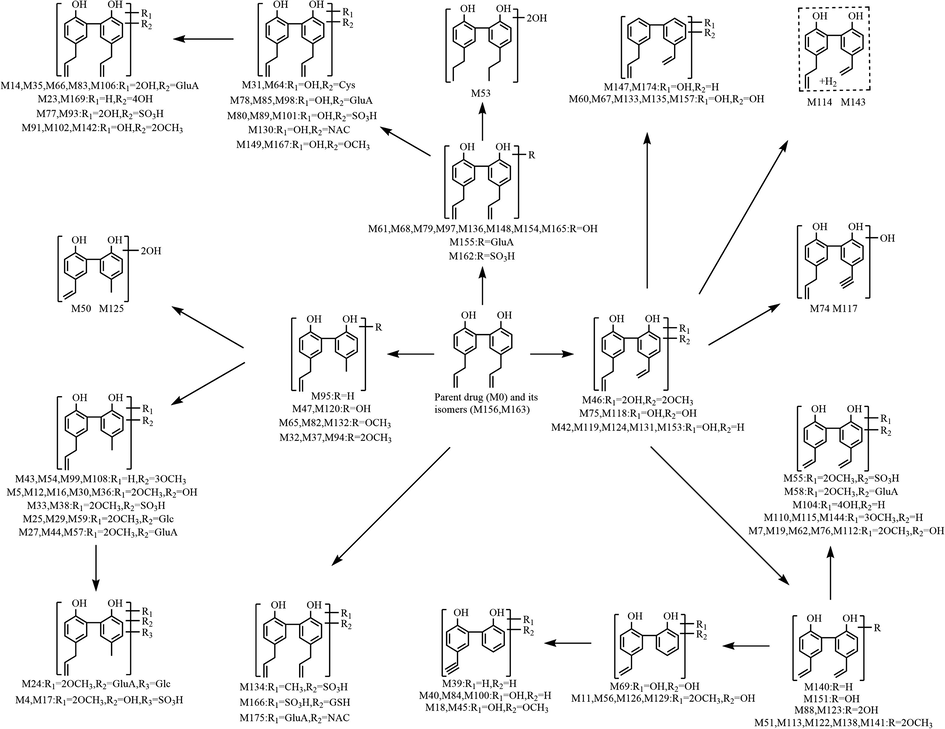
The first metabolic pathway of magnolol in rats in vivo.
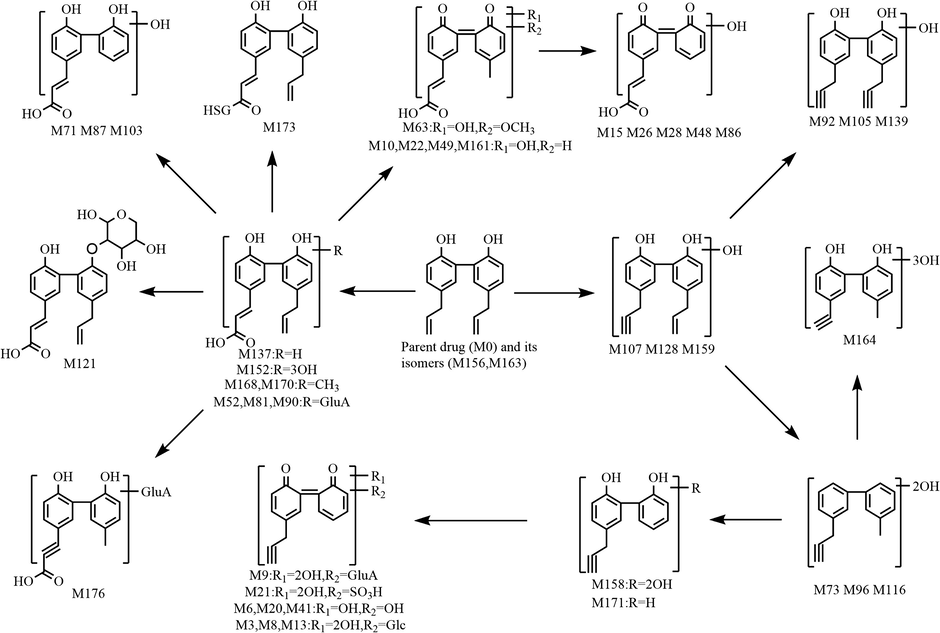
The second metabolic pathway of magnolol in rats in vivo.
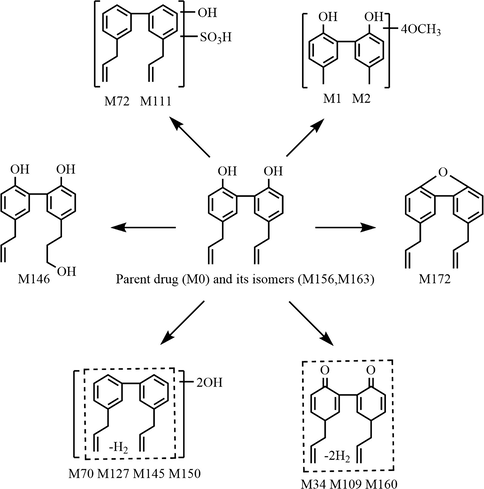
The third metabolic pathway of magnolol in rats in vivo.
4 Conclusion
It is well known that the identification and structural characterization of relevant metabolites is one of the most challenging tasks in the study of drug component metabolism (also known as xenobiotic metabolism), due to the fact that most of the metabolite signals at low concentrations are masked by complex background noise and matrix interference. Hence, a UHPLC-LTQ-Orbitrap MSn-based multi-channel integrated data-mining method was developed to analyze the in vivo metabolism of magnolol in rats, which combined the online data acquisition approach with off-line data processing and structure elucidation techniques. As a result, based on the characteristic fragmentation patterns, chromatographic behaviors, ESI-MS/MS spectral information, and the corresponding Clog P values, a total of 176 metabolites as well as prototype compound were identified in rat urine and plasma, most of which were discovered for the first time.
On the one hand, our results indicated that magnolol originally underwent hydroxylation, methoxylation, oxidation, and carboxylation in rats to generate the corresponding metabolite cluster centers, followed by sulfonation, glucuronidation, glucose conjugation, glutathione conjugation, and N-acetylcysteine conjugation to produce a large number of secondary metabolites. On the other hand, this data-mining strategy based on ultra-high-performance liquid chromatography-high resolution mass spectrometry (UHPLC-HRMS) was validated as a more logical and hierarchical technique for metabolite identification, which could provide a practical reference for future rapid analysis and determination of major-to-trace drug-related components in vivo. In conclusion, the valuable data obtained in our current study not only greatly expanded the understanding of the therapeutic material basis and pharmacological mechanism of magnolol, but also provided new ideas for its toxicity evaluation, safety monitoring and drug delivery forms design.
Author contributions
B. Ma, J. Wang and Z. Wang conceived and designed the research; Y. Guo, Z. Wang and Z. Lin supervised the experimental plan; B. Ma, T. Lou and T. Wang performed UHPLC-LTQ-Orbitrap MS experiment; R. Li, J. Liu, S. Yu, H. Pei, S. Tian and Y. Li performed animal experiment; B. Ma analyzed the data; B. Ma wrote the paper. All authors read and approved the final manuscript.
Acknowledgments
This research was funded by Chinese Pharmacopoeia Commission [grant number, 12] and China National Accreditation Service for Conformity Assessment [grant number, 2020CNAS07].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Benzi, J.R. de L., Moreira, F. de L., Marques, M.P., Duarte, G., Suarez-Kurtz, G., Lanchote, V.L., 2020. A background subtraction approach for determination of endogenous cortisol and 6β-hydroxycortisol in urine by UPLC-MS/MS with application in a within-day variability study in HIV-infected pregnant women. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1144, 122074. https://doi.org/10.1016/j.jchromb.2020.122074.
- The in Vivo Radiosensitizing Effect of Magnolol on Tumor Growth of Hepatocellular Carcinoma. Vivo (Brooklyn).. 2020;34:1789-1796.
- [CrossRef] [Google Scholar]
- Near IR responsive targeted integrated lipid polymer nanoconstruct for enhanced magnolol cytotoxicity in breast cancer. Sci. Rep.. 2020;10:1-12.
- [CrossRef] [Google Scholar]
- The search for, and chemistry and mechanism of, neurotrophic natural products. J. Nat. Med.. 2020;74:648-671.
- [CrossRef] [Google Scholar]
- Cyp3a excipient-based microemulsion prolongs the effect of magnolol on ischemia stroke rats. Pharmaceutics. 2020;12:1-15.
- [CrossRef] [Google Scholar]
- Identification of minor lignans, alkaloids, and phenylpropanoid glycosides in Magnolia officinalis by HPLC-DAD-QTOF-MS/MS. J. Pharm. Biomed. Anal.. 2019;170:153-160.
- [CrossRef] [Google Scholar]
- Suppressive Effects of Hot-water Extract of Magnolia obovata on Clostridium perfringens Enterotoxin-induced Cytotoxicity in Human Intestinal Caco-2 Cells. Planta Med.. 2020;86:198-204.
- [CrossRef] [Google Scholar]
- Magnolia officinalis and Its Honokiol and Magnolol Constituents Inhibit Human Norovirus Surrogates. Foodborne Pathog. Dis. XX. 2020;1–7
- [CrossRef] [Google Scholar]
- Radiosynthesis of a Novel 11C-Labeled Derivative of 4’-O-Methylhonokiol and Its Preliminary Evaluation in an LPS Rat Model of Neuroinflammation. ChemistrySelect. 2020;5:2685-2689.
- [CrossRef] [Google Scholar]
- Magnolol Attenuates Cisplatin-Induced Muscle Wasting by M2c Macrophage Activation. Front. Immunol.. 2020;11:1-17.
- [CrossRef] [Google Scholar]
- Reliable screening and classification of phosphodiesterase type 5 inhibitors in dietary supplements using gas chromatography / mass spectrometry combined with specific common ions. J. Chromatogr. A. 2020;1623:461210
- [CrossRef] [Google Scholar]
- Three unprecedented biphenyl derivatives bearing C6–C3 carbon skeleton from the bark of Magnolia officinalis var. biloba. Chinese Chem. Lett.. 2020;31:1248-1250.
- [CrossRef] [Google Scholar]
- Magnolol prevents acute alcoholic liver damage by activating PI3K/Nrf2/PPARγ and inhibiting NLRP3 signaling pathway. Front. Pharmacol.. 2019;10:1-11.
- [CrossRef] [Google Scholar]
- Comprehensive analysis of resveratrol metabolites in rats using ultra high performance liquid chromatography coupled with high resolution mass spectrometry. Arab. J. Chem.. 2020;13:7055-7065.
- [CrossRef] [Google Scholar]
- Decorative magnolia plants: A comparison of the content of their biologically active components showing antimicrobial effects. Plants. 2020;9:1-9.
- [CrossRef] [Google Scholar]
- Comprehensive metabolism study of polydatin in rat plasma and urine using ultra-high performance liquid chromatography coupled with high-resolution mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci.. 2019;1117:22-35.
- [CrossRef] [Google Scholar]
- Enhanced oral bioavailability of the pharmacologically active lignin magnolol via Zr-based metal organic framework impregnation. Pharmaceutics. 2020;12
- [CrossRef] [Google Scholar]
- Targeted characterization of acylated compounds from Scrophulariae Radix using liquid chromatography coupled with Orbitrap mass spectrometry and diagnostic product ion-based data analysis. J. Sep. Sci.. 2020;43:3391-3398.
- [CrossRef] [Google Scholar]
- A classification of liquid chromatography mass spectrometry techniques for evaluation of chemical composition and quality control of traditional medicines. J. Chromatogr. A. 2020;1609
- [CrossRef] [Google Scholar]
- Ononis spinosa L., an edible and medicinal plant: UHPLC-LTQ-Orbitrap/MS chemical profiling and biological activities of the herbal extract. Food Funct.. 2020;11:7138-7151.
- [CrossRef] [Google Scholar]
- CT1-3, a novel magnolol-sulforaphane hybrid suppresses tumorigenesis through inducing mitochondria-mediated apoptosis and inhibiting epithelial mesenchymal transition. Eur. J. Med. Chem.. 2020;199:112441
- [CrossRef] [Google Scholar]
- Assessment of Biomass Smoke Induced Pulmonary Response and the Protective Effect of Magnolol Using Physiologically Relevant Normal- and Chronic Bronchitis-Like Bronchial Mucosa Models. Care Med: Am. J. Respir. Crit; 2020. p. :201.
- Towards better quality criteria of European honeydew honey: Phenolic profile and antioxidant capacity. Food Chem.. 2019;274:629-641.
- [CrossRef] [Google Scholar]
- Simultaneous determination of honokiol and magnolol in Magnolia officinalis by liquid chromatography with tandem mass spectrometric detection. Biomed. Chromatogr.. 2006;20:1076-1081.
- [CrossRef] [Google Scholar]
- Magnolol Ameliorates Behavioral Impairments and Neuropathology in a Transgenic Mouse Model of Alzheimer’s Disease. Oxid. Med. Cell. Longev.. 2020;2020
- [CrossRef] [Google Scholar]
- Metabolic profiling of coumarins by the combination of UPLC-MS-based metabolomics and multiple mass defect filter. Xenobiotica. 2020;50:1076-1089.
- [CrossRef] [Google Scholar]
- Cardiovascular Modulating Effects of Magnolol and Honokiol, Two Polyphenolic Compounds from Traditional Chinese Medicine-Magnolia Officinalis. Curr. Drug Targets. 2020;21:559-572.
- [CrossRef] [Google Scholar]
- UHPLC-LTQ OrbiTrap MS analysis and biological properties of Origanum vulgare subsp. viridulum obtained by different extraction methods. Ind. Crops Prod.. 2020;154
- [CrossRef] [Google Scholar]
- Profiling and identification of the metabolites of baicalin and study on their tissue distribution in rats by ultra-high-performance liquid chromatography with linear ion trap-Orbitrap mass spectrometer. J Chromatogr. B Anal. Technol. Biomed. Life Sci.. 2015;985:91-102.
- [CrossRef] [Google Scholar]
- Magnolia extract is effective for the chemoprevention of oral cancer through its ability to inhibit mitochondrial respiration at complex i. Cell Commun. Signal.. 2020;18:1-14.
- [CrossRef] [Google Scholar]
- Simultaneous determination of eight bioactive constituents of Zhi-Zi-Hou-Po decoction in rat plasma by ultra high performance liquid chromatography with tandem mass spectrometry and its application to a pharmacokinetic study. J. Sep. Sci.. 2020;43:406-417.
- [CrossRef] [Google Scholar]







