Translate this page into:
Diverse guaiane-type sesquiterpenoids from the root of Daphne genkwa based on molecular networking
⁎Corresponding author. xiaoxiao270@163.com (Xiao-Xiao Huang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Eleven undescribed guaiane-type sesquiterpenoids, named daphnegenols A-K (1–11) and a known analog (12) were targeted for isolation from the root of Daphne genkwa by LC-MS/MS-based molecular networking. Their diverse structures were elucidated through extensive spectroscopic methods, quantum chemical calculation and X-ray single crystal diffraction, including 5/7 bicycle, 5/6/7 tricycles and 5/6/5 tricycles guaiane-type sesquiterpenoid. Daphnegenol A (1) possessed a rare six membered oxyheterocyclic ring which was constructed by oxidation and cyclization at C-1 and C-11. The anti-acetylcholinesterase and anti-neuroinflammatory activities of compounds 1–12 were measured.
Keywords
Daphne genkwa
Guaiane-type sesquiterpenoids
Molecular network
Anti-acetylcholinesterase
Anti-neuroinflammatory
1 Introduction
Guaiane-type sesquiterpenoids are the bicyclic sesquiterpenes possessing a 5/7 fused ring skeleton (Zhou et al., 2018). Oxidation and cyclization reactions could occur at different sites of 5/7 bicycles guaiane-type sesquiterpenoids, and reconstructed into various cyclic systems, affording 5/7/5, 5/6/7, 5/7/6, 5/7/5 tricyclic chemical scaffolds (Zhao et al., 2021). Guaiane-type sesquiterpenoids have attracted increasing attention from chemists and pharmacologists for their wide range of biological activities, including anti-AchE activity (Feng et al., 2014; Zhang et al., 2015), neuroprotective (He et al., 2021; Wang et al., 2020), anti-inflammatory (Kim et al., 2007; Yin et al., 2014; Li et al., 2017), antimalarial (Takaya et al., 1998) and cytotoxic (Park et al., 2006) activity along with their structural diversity.
Guaiane-type sesquiterpenoids were widely distributed in Thymelaeaceae (Ren et al. 2019; Guo et al., 2020), Compositae (Yang et al. 2016), Alismataceae (Peng et al., 2003) and Zingiberaceae, etc (Xiang et al., 2018; Ma et al., 2019). Daphne genkwa Sieb. et Zucc. belong to the Thymelaeaceae family (Hou et al., 2020; Li et al., 2013) and the dried roots have been used in traditional Chinese medicine for a long time to treat edema, breast carbuncle and rheumatism (Hou et al., 2021). The phytochemical studies of the root of D. genkwa revealed the presence of sesquiterpenoids (Feng et al., 2014; Li et al., 2017), biflavonoids (Zheng et al., 2007; Wang et al., 2019a), lignans and coumarins (Baba et al., 1987; Zheng and Shi, 2004). In our prior research on the root of D. genkwa, a series of guaiane-type sesquiterpenoids with potential neuroprotective activity were isolated (Wang et al., 2020; Ren et al., 2019). These are exciting results inspire us to discover more biologically important guaiane-type sesquiterpenoids in polar fraction, and these guaiane-type sesquiterpenoids could guide the separation as the probe compounds in the molecular networking.
Global Natural Products Social Molecular Networking (GNPS) is an open accessed platform, which was used to analyze the LC-MS/MS data of extracts and feedback the molecular networking of the selected samples and probe compound (Wang et al., 2016). The probe compound will cluster with similar compounds in the molecular networking, and based on the chromatographic information, the guided separation of the nodes was achieved (Xu et al., 2022a). With the guides of molecular networking, 12 guaiane type sesquiterpenoids were isolated from the root of D.genkwa. Among of them, compounds 1–11 were undescribed compounds. The chemical structures of undescribed compounds were unequivocally identified by detailed 1D and 2D NMR, HRESIMS, NMR calculations, ECD and X-ray diffraction. Structurally, daphnegenol A (1) possessed a rare six membered oxyheterocyclic ring which were constructed by oxidation and cyclization at C-1 and C-11. In addition, the anti-AchE and anti-neuroinflammatory activities of compounds 1–12 were examined.
2 Materials and methods
2.1 General experimental procedures
Optical rotations were obtained on an Anton Paar MCP200 digital polarimeter (Anton Paar, Bremen, Germany). UV spectra were measured on a Shimadzu UV-2600i spectrometer (Shimadzu, Kyoto, Japan). ECD experiments were recorded on a MOS 450 spectrometer (Bio-logic, seyssinet Pariset, France). The high-resolution electrospray ionization mass spectrometry (HRESIMS) data were performed on a Micro Q-TOF spectrometer (Bruker Daltonics, Billerica, USA). The NMR spectra were obtained on Bruker AVAVCE III HD 600 MHz (Bruker BioSpin, Flanden, Switzerland) with TMS as an internal standard. The X-ray single crystal diffraction data were obtained on D8 QUEST single crystal diffractometer (Bruker AXS, Bremen, Germany). The melting point was obtained with an X-6 melting-point apparatus (Beijing TECH Instrument Co. ltd, China). Semi-preparative RP-HPLC separations were performed on instruments equipped with a LC-6AD high-pressure pump with an SPD-20A ultraviolet–visible (UV–vis) light absorbance detector using a YMC C18 semi-preparative column (250 mm × 10 mm, 5 μm, Shimadzu, Tokyo, Japan). Column chromatography were performed on silica gel (100–200 or 200–300 mesh, Qingdao Marine Chemical Co. ltd., Qingdao, People’s Republic of China), HP20 macroporous resin (75–150 μm, Tokyo, Japan), polyamide resin (100 mesh, Qingdao, People’s Republic of China) and octadecyl silica gel (ODS) (60–80 μm, YMC Co. ltd., Kyoto, Japan).
BV-2 cell (ATCC, Maryland, USA) cultured in carbon dioxide incubator (Thermo, Massachusetts, USA). The OD values were collected on a Varioskan Flash (Thermo, Massachusetts, USA), other reagent include acetylcholinesterase (AchE), acetylthiocholine iodide (ACTI), 5,5′-dithiobis(2-nitrobenzoic acid) (DNTB) and positive control donepezil were purchased from Shanghai Macklin Biochemical Co. ltd, China. Greiss reagent (Beyotime Biotech, Shanghai, China), Dulbecco’s modified Eagle’s medium (DMEM, Hyclone, Utah, USA), fetal bovine serum (FBS, Hyclone, Utah, USA), antibiotics (Hyclone, USA) and LPS (sigma, USA) were used in Nitric oxide (NO) production assay.
2.2 Plant material
The dried roots of D. genkwa were collected from Xingyi City (104°53′E, 24°80′N), Guizhou Province, People’s Republic of China, in July 2017, and were identified by Prof. Jin-Cai Lu (Department of Traditional Chinese Materia Medica, Shenyang Pharmaceutical University, People’s Republic of China). A voucher specimen (No. 20170721) has been deposited in the Department of Natural Products Chemistry, Shenyang Pharmaceutical University for further reference.
2.3 Extraction and isolation
The root of D. genkwa (50 kg) were exhaustively extracted with 70 % EtOH for three times under reflux. The extract (4.1 kg) was partitioned successively with PE, EtOAc and n-BuOH. The EtOAc extract (990 g) was separated by silica gel column chromatography involving elution with PE-EtOAc and CH2Cl2-MeOH gradient system to give four fractions A-D. Fraction C (80 g) was subjected to the polyamide column chromatography involving elution with EtOH-H2O (20:80 and 80:20, v/v) to yield two fractions C1 and C2. Fraction C1 (12 g) was purified by ODS silica gel using EtOH-H2O, and further separated by silica gel column chromatography involving elution with CH2Cl2-MeOH gradient system (50:1 → 3:1) to give six subfractions Fr. C1-1 to Fr. C1-6. Compounds 1–12 were separated by HPLC with MeOH-H2O (10:90 to 40:60, v/v) and MeCN-H2O (10:90 to 15:85, v/v) gradient system. Compounds 1 (82.9 mg, tR = 25.4 min), 2 (23.6 mg, tR = 27.8 min), 4 (8.6 mg, tR = 34.9 min) and 5 (2.9 mg, tR = 40.7 min) were separated from subfraction Fr. D1-5 with MeCN-H2O (15:85, v/v, 3.0 mL/min) gradient system. Compounds 3 (6.7 mg, tR = 28.1 min), 6 (4.0 mg, tR = 33.0 min), 10 (5.0 mg, tR = 35.8 min) were obtained from subfraction Fr. C1-2. In addition, compounds 7 (23.5 mg, tR = 14.7 min), 8 (31.9 mg, tR = 19.5 min) and 9 (6.2 mg, tR = 23.1 min) were separated from subfraction Fr. C1-3 with MeCN-H2O (10:90, v/v, 3.0 mL/min) gradient system. Subfractions Fr. C1-6 and Fr. C1-1 produce compound 11 (3.8 mg, tR = 30.8 min) and compound 12 (12.4 mg, tR = 36.4 min) with MeCN-H2O (10:90, v/v, 3.0 mL/min) gradient system.
Daphnegenol A (1): colorless crystal (MeOH); mp: 162–164 °C; [α] −62.7 (c 0.10 MeOH); UV (MeOH) λmax (log ε) 232 (2.80) nm; 1H NMR data (DMSO‑d6, 600 MHz) see Table 1, 13C NMR data (DMSO‑d6, 150 MHz) see Table 3, and HRESIMS m/z 289.1415 [M+Na]+, calcd for 289.1410.
No.
1
2
3
4
5
6
2
2.70, d (16.6)
1.85, d (16.6)2.13, m
1.40, overlap
5.64, s
5.64, d (1.8)
3
1.68, m
1.63, overlap2.47, dd (18.2, 6.5)
1.87, dd (18.2, 2.2)
2.45, dd (16.4, 7.7)
1.95, br. d (16.4)
4
2.60, m
2.41(dq, 7.5, 7.8)
2.03, m
2.14, m
5
1.83, m
3.37, m
2.77, m
3.82, m
6
2.75, m
2.50, overlap1.64, overlap
1.51, dd (12.7, 10.1)2.67, d (18.9)
2.28, d (18.9)1.87, dd (12.1, 8.7)
1.49, dd (12.1, 12.1)2.24, o
1.40, o1.73, ddd (14.3, 10.8, 3.1)
1.45, ddd (14.3, 13.0, 6.1)
7
2.15, m
2.08, overlap
8
1.87, overlap
1.55, m
4.20, ddd (10.2, 4.6, 3.2)
4.32, br. d (10.8)
4.33, dd (10.8, 3.7)
4.46, overlap
9
1.47, m
5.96, d (1.2)
2.14, dd (13.3, 10.2)
1.29, dd (13.3, 3.2)2.24, dd (14.1, 10.8)
1.44, dd (14.1, 4.2)2.23, overlap
1.39, overlap4.06, d (5.5)
10
11
1.98, m
12
1.08, s
1.92, m
1.41, overlap0.83, s
0.66, s
0.67, s
3.32, overlap
3.14, d (12.1)
13
3.56, dd (10.5, 5.2)
3.11, dd (10.5, 5.2)3.32, overlap
2.91, m3.79, dd (11.1, 2.1)
3.46, dd (11.1, 5.6)3.78, dd (11.1, 2.3)
3.45, dd (11.1, 5.5)3.79, d (11.1)
3.44, d (11.1)3.31, overlap
3.26, dd (11.4, 5.5)
14
1.14, s
2.02, d (1.2)
1.16, s
1.04, s
1.03, s
2.09, s
15
1.62, s
1.17, s
1.07, d (7.1)
0.96, d (7.8)
1.04, overlap
0.72, d (7.1)
OH-4
4.38, overlap
OH-7
4.77, s
5.18, s
5.01, s
4.99, s
OH-8
4.73, d (4.6)
5.05, d (3.8)
4.99, overlap
5.10, d (4.6)
OH-10
4.40, s
OH-11
4.38, s
OH-13
4.73, t (5.2)
4.37, overlap
4.71, dd (5.6, 2.1)
4.80, br. s
4.80, br. s
4.46, overlap
No.
7
8
9
10
11
2
2.42, d (17.7)2.28, d
(17.7)2.89, d (18.5)
2.13, d (18.5)2.53, d (17.7)
2.14, d (17.7)
2.71, d (17.4)
2.10, d (17.4)
3
3.91, d (6.2)
4
5
6
2.65, m
2.32, overlap2.65, br. d (19.0)
2.51, overlap2.69, dd (11.9, 11.6)
2.41, dd (11.6, 2.0)2.61, br. d (15.5)
2.42, dd (15.5,11.5)2.99, d (15.5)
2.46, d (15.5)
7
2.32, overlap
2.84, m
2.00, m
1.84, overlap
2.99, overlap
8
1.78, br. d (14.2)
0.95, overlap1.65, br. d (14.3)
1.29, m1.85, m
1.53, ddd (13.9, 8.1, 2.9)1.85, overlap
1.71, overlap3.98, m
9
1.98, m
1.35, m2.20, m
1.45, m1.90, ddd (14.4, 8.1, 3.2)
1.32, ddd (14.4, 9.7, 2.9)1.70, overlap
1.37, m2.09, dd (14.5, 1.7)
1.82, dd (14.5, 5.5)
10
2.10, m
2.83, m
11
12
0.95, s
4.96, d (1.6)
4.84, d (1.6)4.98, d (1.6)
4.86, d (1.6)4.99, d (1.6)
4.89, d (1.6)5.00, s
13
3.30, s
3.93, m
3.95, d (5.5)
3.96, d (5.0)
3.95, m
14
0.61, d (7.0)
0.82, s
1.05, s
0.97, d (7.3)
1.17, s
15
1.56, s
1.53, s
1.58, s
1.07, s
1.60, s
OH-1
5.20, s
5.25, s
5.12, s
5.47, s
OH-3
5.60, d (6.2)
OH-4
5.23, s
OH-8
5.28, d (5.6)
OH-10
4.45, s
4.56, s
4.70, s
OH-11
3.99, s
OH-13
4.42, br. s
4.80, t (5.5)
4.82, t (5.5)
4.86, t (5.0)
4.90, t (5.4)
Position
1
2
3
4
5
6
7
8
9
10
11
1
83.9
50.9
143.6
190.7
189.4
138.0
81.2
84.4
80.7
141.2
83.8
2
47.3
32.2
204.9
122.4
122.6
207.7
50.7
48.1
46.6
202.2
47.8
3
203.9
39.9
44.7
211.3
209.4
48.7
204.3
205.2
206.1
81.1
205.1
4
136.2
78.6
33.7
42.6
49.1
30.6
136.3
136.4
135.5
77.9
136.5
5
166.9
56.9
176.5
38.3
43.7
39.4
173.2
171.5
173.6
171.8
171.8
6
24.6
33.6
33.9
30.8
35.3
29.4
30.6
36.4
30.4
32.5
29.3
7
32.1
81.0
83.5
83.7
83.0
39.1
41.2
36.9
41.6
41.2
41.5
8
22.3
202.1
76.8
75.1
75.0
67.2
23.6
31.3
27.9
30.6
71.6
9
36.0
127.5
46.6
42.9
42.7
81.1
31.0
39.4
34.9
32.2
44.2
10
75.5
172.7
43.6
46.4
46.1
143.0
39.2
75.2
72.7
25.9
76.1
11
76.6
45.0
49.2
50.2
50.3
75.0
73.9
155.9
155.0
155.4
153.4
12
24.2
30.5
12.3
13.0
13.4
62.4
20.3
106.5
106.7
106.8
108.2
13
66.1
62.0
64.2
64.5
64.6
64.2
67.6
63.0
62.8
63.0
63.3
14
24.8
22.3
14.1
17.1
17.0
18.5
14.4
22.5
25.4
16.6
27.3
15
7.8
26.3
18.2
12.6
13.6
15.6
7.6
7.5
7.2
22.4
7.8
Daphnegenol B (2): colorless syrup (MeOH); [α] + 29.6 (c 0.10 MeOH); UV (MeOH) λmax (log ε) 244 (2.74) nm; 1H NMR data (DMSO‑d6, 600 MHz) see Table 1, 13C NMR data (DMSO‑d6, 150 MHz) see Table 3, and HRESIMS m/z 289.1412 [M+Na]+, calcd for 289.1410.
Daphnegenol C (3): colorless crystal (MeOH); mp: 161–163 °C; [α] −49.1 (c 0.10 MeOH); UV (MeOH) λmax (log ε) 232 (2.74) nm; 1H NMR data (DMSO‑d6, 600 MHz) see Table 1, 13C NMR data (DMSO‑d6, 150 MHz) see Table 3, and HRESIMS m/z 267.1587 [M+H]+, calcd for 267.1591.
Daphnegenol D (4): colorless syrup (MeOH); [α] −40.1 (c 0.10 MeOH); UV (MeOH) λmax (log ε) 234 (2.89) nm; 1H NMR data (DMSO‑d6, 600 MHz) see Table 1, 13C NMR data (DMSO‑d6, 150 MHz) see Table 3, and HRESIMS m/z 289.1412 [M+Na]+, calcd for 289.1410.
Daphnegenol E (5): colorless syrup (MeOH); [α] −39.8 (c 0.10 MeOH); UV (MeOH) λmax (log ε) 233 (2.62) nm; 1H NMR data (DMSO‑d6, 600 MHz) see Table 1, 13C NMR data (DMSO‑d6, 150 MHz) see Table 3, and HRESIMS m/z 289.1411 [M+Na]+, calcd for 289.1410.
Daphnegenol F (6): colorless syrup (MeOH); [α] −44.5 (c 0.10 MeOH); UV (MeOH) λmax (log ε) 252 (2.54) nm; 1H NMR data (DMSO‑d6, 600 MHz) see Table 1, 13C NMR data (DMSO‑d6, 150 MHz) see Table 3, and HRESIMS m/z 305.1346 [M+Na]+, calcd for 305.1359.
Daphnegenol G (7): colorless syrup (MeOH); [α] + 74.8 (c 0.10 MeOH); UV (MeOH) λmax (log ε) 240 (2.75) nm; 1H NMR data (DMSO‑d6, 600 MHz) see Table 2, 13C NMR data (DMSO‑d6, 150 MHz) see Table 3, and HRESIMS m/z 291.1555 [M+Na]+, calcd for 291.1567.
Daphnegenol H (8): colorless syrup (MeOH); [α] + 44.4 (c 0.10 MeOH); UV (MeOH) λmax (log ε) 242 (2.75) nm; 1H NMR data (DMSO‑d6, 600 MHz) see Table 2, 13C NMR data (DMSO‑d6, 150 MHz) see Table 3, and HRESIMS m/z 289.1415 [M+Na]+, calcd for 289.1410.
Daphnegenol I (9): colorless syrup (MeOH); [α] −50.0 (c 0.10 MeOH); UV (MeOH) λmax (log ε) 239 (2.84) nm; 1H NMR data (DMSO‑d6, 600 MHz) see Table 2, 13C NMR data (DMSO‑d6, 150 MHz) see Table 3, and HRESIMS m/z 289.1400 [M+Na]+, calcd for 289.1410.
Daphnegenol J (10): colorless syrup (MeOH); [α] + 22.2 (c 0.10 MeOH); UV (MeOH) λmax (log ε) 240 (2.74) nm; 1H NMR data (DMSO‑d6, 600 MHz) see Table 2, 13C NMR data (DMSO‑d6, 150 MHz) see Table 3, and HRESIMS m/z 289.1407 [M+Na]+, calcd for 289.1410.
Daphnegenol K (11): colorless syrup (MeOH); [α] −56.7 (c 0.10 MeOH); UV (MeOH) λmax (log ε) 237 (2.85) nm; 1H NMR data (DMSO‑d6, 600 MHz) see Table 2, 13C NMR data (DMSO‑d6, 150 MHz) see Table 3, and HRESIMS m/z 289.1415 [M+Na]+, calcd for 289.1410.
2.4 X-ray crystallographic analysis of 1, 3 and 12
Colorless single crystal of compounds 1, 3 and 12 were obtained by recrystallization from MeOH. All X-ray crystallographic data were acquired by a Bruker D8 QUEST diffractometer with Cu Kα (radiation λ = 1.54178 Å) at T = 153 K. Their structures were solved by SHELXS method and then refined anisotropically with SHELXL-2018/3 using a full-matrix least-squares procedure based on F2. The crystallographic data of 1, 3, and 12 have been deposied with the Cambridge Crystallographic Data Centre (CCDC number 2158257, 2158256, 2158258). The data can be obtained, free of charge, from the Cambridge Crystallographic Data Centre (https://www.ccdc.cam.ac.uk/data_request/cif).
Crystallographic data of 1: colorless crystal, crystal size 0.100 × 0.200 × 0.200 mm3, C15H22O4, M = 266.32, orthorhombic space group P21 21 21, a = 7.1287(3) Å, b = 8.5317(3) Å, c = 21.7552(8) Å, V = 1323.15(9) Å3, Z = 4, Dcalcd = 1.337 g/cm3, 9111 collected reflections (8.128◦ ≤ 2θ ≤ 136.4◦), μ (Cu Kα) = 0.780 mm−1, R1 = 0.0271 and wR2 = 0.0696 for I ≥ 2σ (I), S = 1.041, absolute structure parameter (Flack parameter) = 0.06 (5).
Crystallographic data of 3: colorless crystal, crystal size 0.100 × 0.200 × 0.300 mm3, C15H22O4, M = 266.32, monoclinic space group P1 21 1, a = 7.0304(5) Å, b = 9.3576(7) Å, c = 10.4322(8) Å, α = 90°, β = 91.106(2)°, γ = 90°, V = 686.18(9) Å3, Z = 2, Dcalcd = 1.289 g/cm3, 9358 collected reflections (8.477◦ ≤ 2θ ≤ 144.7◦), μ (Cu Kα) = 0.752 mm−1, R1 = 0.0471 and wR2 = 0.1318 for I ≥ 2σ (I), S = 1.064, absolute structure parameter (Flack parameter) = −0.03 (8).
Crystallographic data of 12: colorless crystal, crystal size 0.100 × 0.100 × 0.200 mm3, C15H20O4, M = 264.31, triclinic space group P1, a = 7.0186(7) Å, b = 7.1490(7) Å, c = 7.1885(7) Å,α = 73.650(4)°, β = 72.873(5)°, γ = 83.555(4)°, V = 33.0.59(6) Å3, Z = 1, Dcalcd = 1.328 g/cm3, 3438 collected reflections (12.91◦ ≤ 2θ ≤ 149.0◦), μ (Cu Kα) = 0.780 mm−1, R1 = 0.0516 and wR2 = 0.1484 for I ≥ 2σ (I), S = 1.110, absolute structure parameter (Flack parameter) = 0.04 (19).
2.5 LC–MS/MS and molecular networking analysis
The fractions from D. genkwa were prepared and tested by the AB SCIEX 4000 Q-Trap MS Instrument. The ESI source, EMS-IDA-EPI mode, and positive ion mode were employed. The analysis condition from 10:90 to 50:50 (MeCN: H2O) in 50 min was used. MS scans were operated from m/z 0–1200. MSConvert software was applied to the.d format of MS/MS data to the.mzML format, then the file was upload to GNPS (Wang et al., 2016) by WinScp 5.19.5. Finally, a molecular network was created using the GNPS Web platform (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=856717c3e18a47d3a5fc5bfd084f2482). The network analysis was visualized by Cytoscape 3.9.2 software.
2.6 ECD and NMR calculations
Conformational searches of 1–11 were performed by Spartan software in the MMFF force field. After geometry optimization by DFT calculations, the conformers which Boltzmann Distribution above 0.1 % were selected. Next, these conformations were further optimized by the density functional theory method at the B3LYP/6-31G(d) level in Gaussian 09 program package (Frisch et al., 2016). The NMR calculations were performed using gauge independent atomic orbital (GIAO) method at the mPW1PW91/6-311+G(d,p) levels in DMSO solution. Boltzmann-weighted averages of the chemical shifts were calculated by shermo (Lu and Chen, 2021) to scale them against the experimental values. The experimental and calculated data were analyzed by the improved probability DP4+ method for isomeric compounds (Wang et al., 2019b).
The absolute configurations were determined by using time-dependent density functional theory (TDDFT) calculations. The theoretical ECD calculations were performed using TDDFT method at the B3LYP/6-311++G (2d, p) levels in MeOH solution. The ECD curves were generated by SpecDis 1.51 software on the basis of the Boltzmann weighting of each conformer.
2.7 Bioassay for the inhibition of acetylcholinesterase (AchE)
The bioassay for the inhibition of AchE were valued by Ellman method (Al Muqarrabun et al., 2014). ACTI was used as substrate in the assay. Compounds were dissolved in DMSO. The mixture contained 50 μL phosphate buffer (pH 8.0), 25 μL of compound at 10 μM and 100 μM, 125 μL of DTNB (0.5 mM) and 12.5 μL AChE (0.1 U/mL), and refrigerated the mixture at 4 ℃ overnight. The reaction was initiated by the addition of 50 μL of ACTI (0.15 mM). The hydrolysis of ATCI was monitored at 412 nm after 5 min. Donepezil was used as positive control. All reactions were performed in triplicate.
2.8 Molecular docking
Crystal structure of human AChE (PDB code:1EVE) was obtained from RCSB protein Data Bank. The lowest energy conformations of the compounds 6 and 11 were calculated with LigPrep, and the protein structures were pre-processed, refined, and corrected using the Protein Preparation Wizard module. Afterward, for the dockings, the grid center was placed on the centroid of the included ligand binding site. Finally, the molecular docking results were processed by PyMOL (Xu et al, 2022b).
2.9 Bioassay for the inhibition of NO production
Murine microglial cells BV-2 were cultured in DMEM supplemented with 10 % (v/v) FBS and 1 % antibiotics containing 5 % CO2 at 37 °C. The bioassay for compounds to inhibit NO production was conducted by the method in the literature (Guo et al., 2021; Bai et al., 2022). BV-2 microglia cells were seeded in 96-well plates and then treated with or without the compound at 10 μM for 1 h. In each well, LPS (10 μg/mL) was added to induce inflammation then culture for 24 h. The NO production in the supernatant was quantified by the Griess reaction. The absorbance at 540 nm was measured in a multifunctional enzyme marker. The NO concentration was calculated via GraphPad Prism.
3 Results and discussion
The EtOAc partition of the root of D. genkwa was fractionated over a silica gel chromatography column to yield four fractions A-D. In our previous study (Wang et al., 2019b, 2020; Ren et al., 2019; Guo et al., 2020), Fr. A and B were separated systematically to obtain guaiane-type sesquiterpenoids and other types of secondary metabolites, which would reduce separation efficiency. To guided separated guaiane-type sesquiterpenoid from Fr. C1, daphnegen D was analyzed as a reference probe to construct initial focal points followed by comparison and organization with the MS/MS data of Fr. C1, in which compounds with similar cracking laws form nodes. The LC-MS/MS data of Fr. C1 and the daphnegen D were collected, and these data were upload to the GNPS web platform, and similar spectra were clustered as molecular families. Molecular network analysis revealed that some nodes clustered with the reference probe (daphnegen D, m/z 267.0), which were considered as guaiane-type sesquiterpenoids or its derivatives according to the similarity of cosine value (Xu et al., 2022a) (see Fig. 1).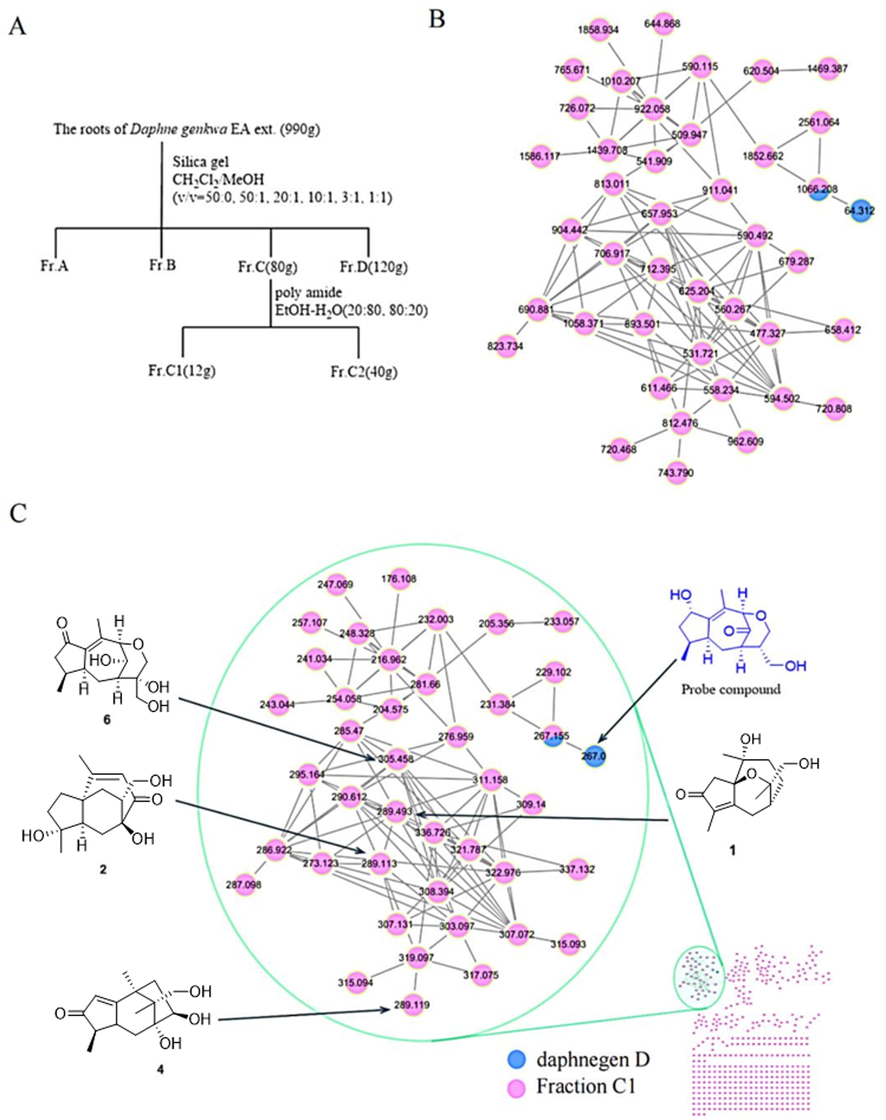
A) The separation flow chart of the root of D. genkwa. B) The molecular network with RTmean label was established by LC-MS/MS analysis of fraction C1 in D. genkwa (pink nodes) and daphnegen D (blue nodes). C) The molecular network with parent mass label was established by LC-MS/MS analysis of fraction C1 in D. genkwa (pink nodes) and daphnegen D (blue nodes).
To further analyzed the LC-MS/MS data of these nodes clustered with probe reference, the parent mass was employed as the important evidence to differentiated sesquiterpenoids from others (Mi et al., 2022). Previous studies showed that the root of D. genkwa also exist lignans, and its molecular weight were above 400, while the molecular weight of sesquiterqenoids were between 200 and 300. The molecular network displayed a cluster with the reference probe, and the parent mass of nodes in the cluster mostly between 200 and 350, which is accorded with the structural characteristics of sesquiterpenoids. The molecular network displayed that the RTmean of sesquiterqenoids mostly among 400 to 1000 s, while RTmean of lignans mostly were above of 2000 s, which were served as the important basis for guiding separation. Therefore, Fr. C1 was further separated by a silica gel chromatography column and give six subfractions Fr. C1-1 to Fr. C1-6. Finally, compounds 1, 2, 4, 5 were separated from Fr. C1-5, compounds 3, 6, 10 from Fr. C1-2, and compounds 7–9 from Fr. C1-3. Others were separated from Fr. C1-6 and Fr. C1-1 (see Fig. 2).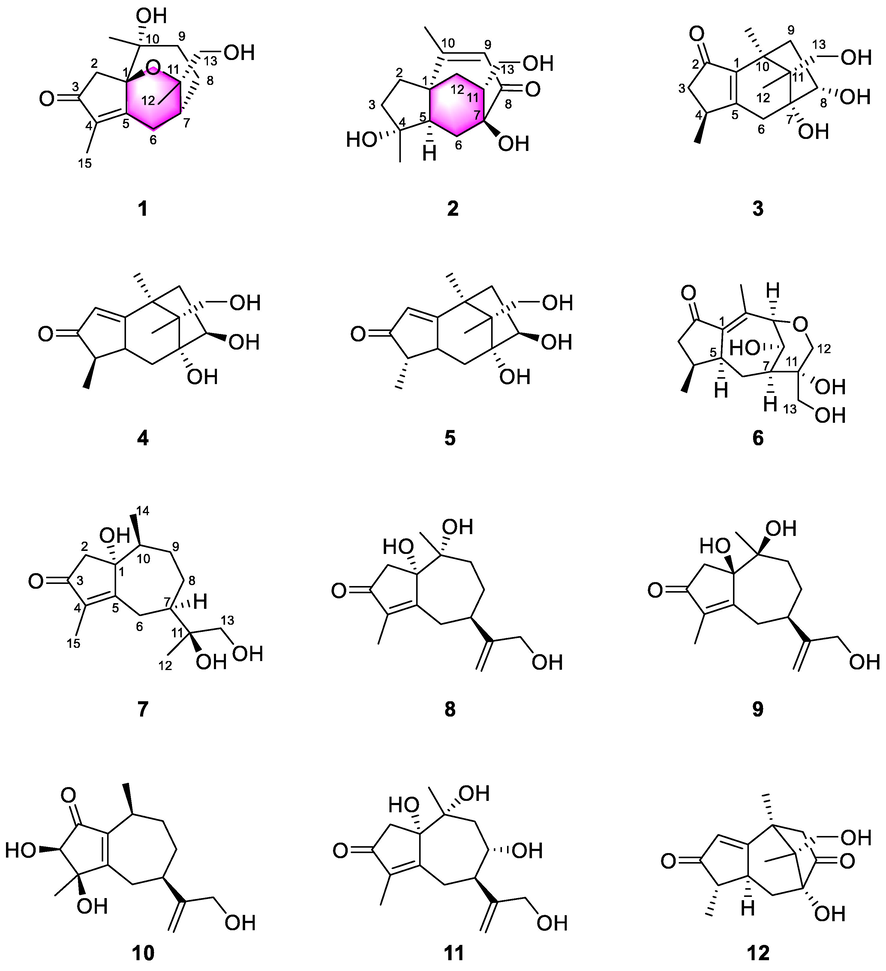
Chemical structures of compounds 1–12.
Compound 1 was obtained as colorless crystal, and the molecular formula was assigned as C15H22O4 via the HRESIMS (m/z 289.1415 [M+Na]+, calcd for 289.1410) data, requiring 5 degrees of unsaturation. The 1H NMR data of 1 exhibited three methyl proton signals at δH 1.62 (3H, s, H-15), 1.14 (3H, s, H-14), 1.08 (3H, s, H-12), a methine signal at δH 2.15 (1H, m, H-7) and nine methene signals at δH 2.70 (1H, d, J = 16.6 Hz, H-2α), 1.85 (1H, d, J = 16.6 Hz, H-2β), 2.75(1H, dd, J = 19.5, 5.6 Hz, H-6β), 2.50 (1H, overlap, H-6α), 1.87(1H, overlap, H-8β), 1.55(1H, m, H-8α), 1.47(2H, m, H-9), 3.56(1H, dd, J = 10.5, 4.5 Hz, H-13a), 3.11(1H, dd, J = 10.5, 4.8 Hz, H-13b). The 13C NMR spectrum displayed 15 carbon resonances, including one carbonyl carbon at δC 203.9, two olefinic carbons at δC 166.9, 136.2, four oxygenated carbons at δC 83.9, 75.5, 76.6, 66.1 and eight aliphatic carbons at δC 47.3, 24.6, 32.1, 22.3, 36.0, 24.2, 24.8, 7.8. Furthermore, δC 203.9, 166.9 and 136.2 illustrated the presence of α, β-unsaturated ketone.
Combined with HSQC data, twelve aliphatic carbons were identified as three quaternary carbons, one tertiary carbon, five secondary carbons and three primary carbons. δH 4.73 (1H, t, J = 5.2 Hz, OH-13), 4.40 (1H, s, OH-10) were identified as exchangeable proton signals according to HSQC data. Analysis of 1H-1H COSY spectrum displayed a key spin-coupled systems [CH2(6)-CH(7)-CH2(8)]. The key HMBC correlations of H-2/C-1, C-3, C-4, C-5, H-7/C-5, C-9, H-9/C-7, C-1, H-6/C-1 and H-8/C-10 revealed the existence of 5/7 fused ring skeleton. The correlation between H-14 and C-3, C-4, C-5 supported the existence of α, β- unsaturated ketone and specified the position of carbonyl and double bond. Also, key HMBC correlations of H-12/C-11, C-7, H-14/C-1, C-9 and H-15/C-3, C-5 displayed three methyl were substituted at C-11, C-10 and C-4, the correlations of H-12/C-7 and H-13/C-7 illustrated that C-11 was linked to C-7. Thus, the gross structure of 1 was identified as a guaiane-type sesquiterpenoid. The groups mentioned above occupied four degrees of unsaturation, besides, an oxygen bridge is formed to satisfy the remaining unsaturation. The 13C NMR data showed C-1, C-11, C-10 and C-12 were assigned as oxygenated carbons, but only have HMBC correlations of OH-13/C-12 and C-11, OH-10/C-10, C-1 and C-9. It also supported the existence of six membered oxyheterocyclic ring (Yin et al. 2014) (see Fig. 3).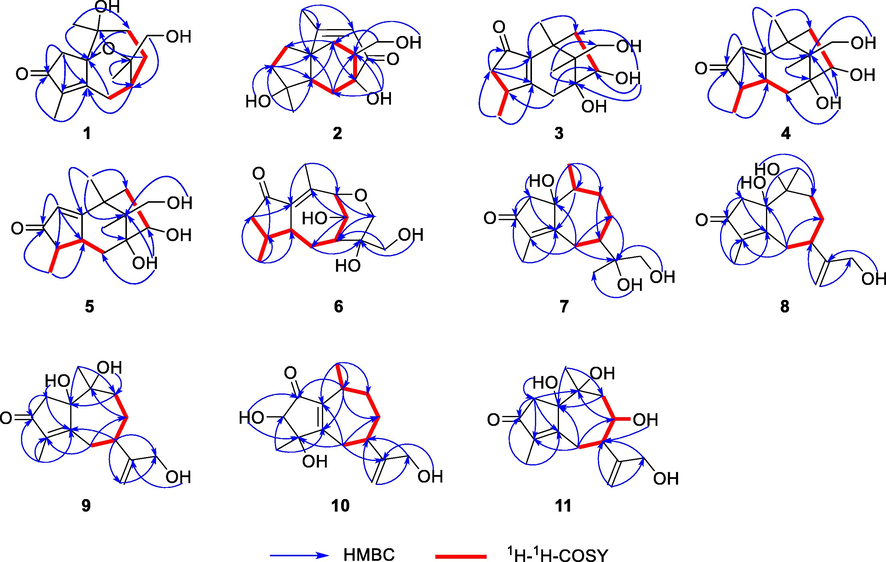
Key HMBC (blue) and 1H−1H COSY (bold red) correlations of compounds 1–11.
The NOESY correlations of H-6β (δH 2.75)/H-12 (δH 1.08) and H-14 (δH 1.14)/H-13a (δH 3.56) displayed that H-6β and H-12 were at homo side, and H-14 and H-13a were at homo side. However, the relative configuration could not be completely identified by NOESY spectrum for the deficiency of hydrogen at C-1 and C-10 (see Fig. 4). Fortunately, the single crystal of 1 was obtained and the relative and absolute configuration of 1 was unambiguously assigned (see Fig. 5). The experimental ECD curve of 1 resembled the calculated ECD curve for (1R,7R,10R,11S)-1, which further supported absolute configuration of 1 (see Fig. 7). The structure of 1 was confirmed as shown in Fig. 2 and was named as daphnegenol A, which was identified as a guaiane type sesquiterpenoid with a six membered oxyheterocyclic ring.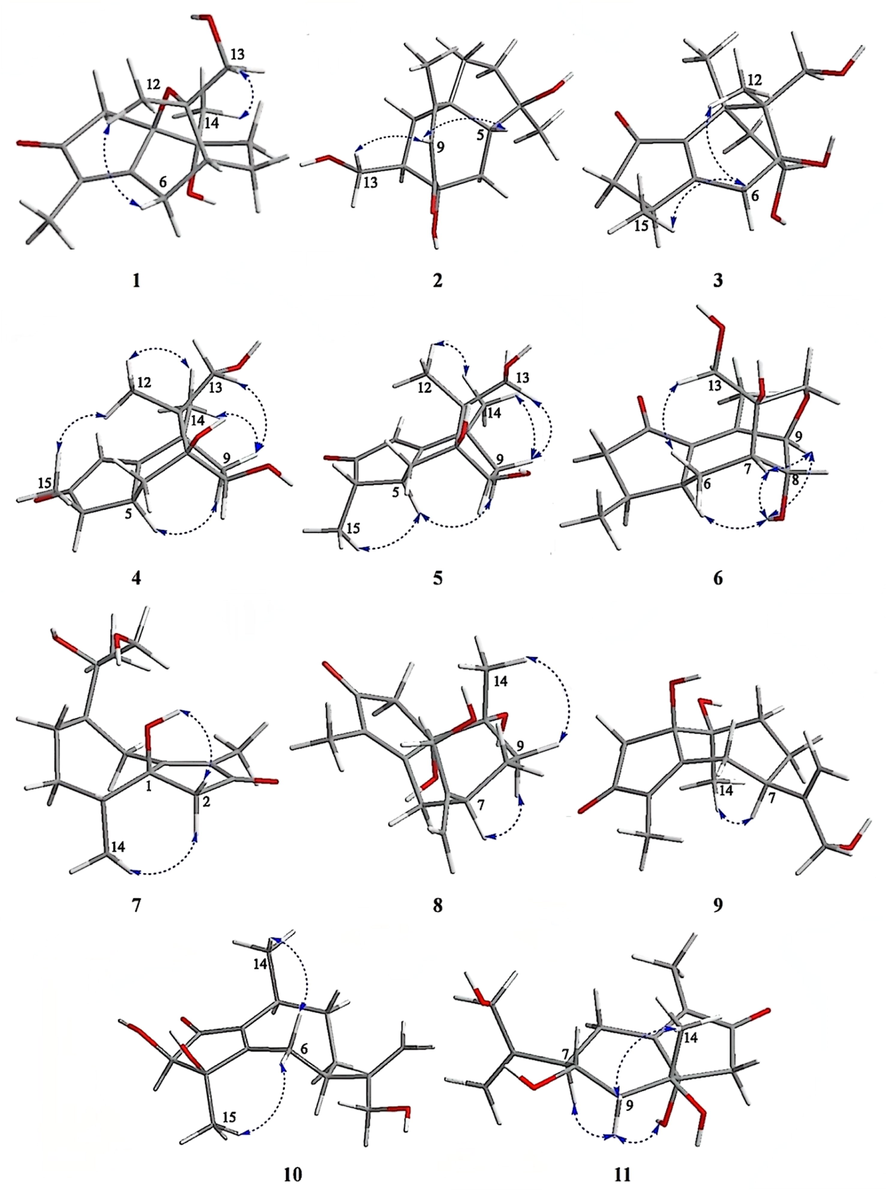
The key NOESY correlations of compounds 1–11.
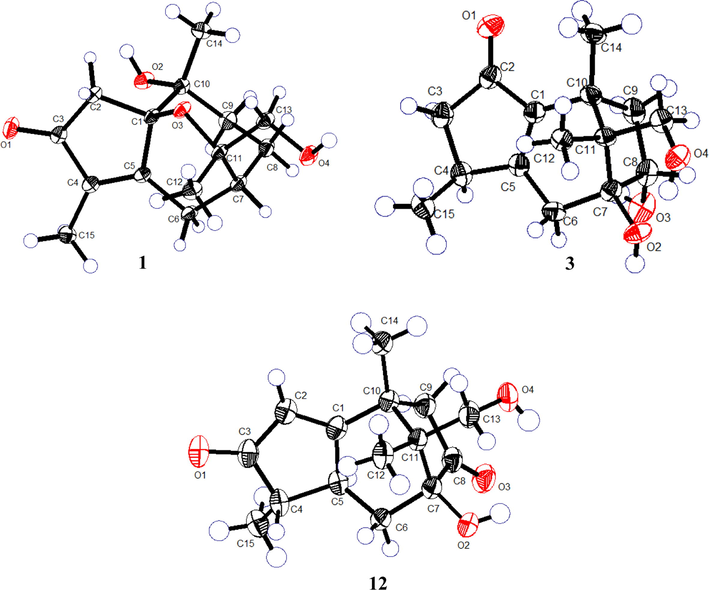
The ORTEP drawing of compounds 1, 3 and 12.
Daphnegenol B (2) was found to possess the molecular formula of C15H22O4, according to the positive ion at m/z 289.1414 [M+Na]+ (calcd for 289.1410) in the HRESIMS data. Compound 2 was deduced to be an analog of daphnauranol A (Huang et al., 2014), except that the signals of an aliphatic carbon C-4 (δC 34.9) in the 13C NMR spectrum of daphnauranol A was replaced by an oxygenated carbon (δC 78.6). Along with the HMBC correlations of OH-4/C-3, C-4 and C-5 showed that compound 2 was the hydroxy substitution of daphnauranol A. According to HMBC correlations of H-5/C-12, H-12/C-10 and H-12/C-1, C-12 was linked to C-1 and formed a six-member-ring. The gross structure was identified as a rare 5/6/7 tricycles guaiane type sesquiterpene only four compounds were reported (Huang et al., 2014; Ma et al., 2017).
The NOESY spectrum showed correlations of H-5 (δH 1.83)/H-9 (δH 5.96), H-9 (δH 5.96)/H-13 (δH 3.32, 2.91), which confirmed that the carbon bridge C-7/C-8/C-9/C-10/C-1, H-5 and H-13 were below the molecular. The relative configuration of C-4 was not identified for the signal overlap of H-3 and H-6. To assignment the relative configuration of compound 2, the calculated NMR of two possible conformers (1R*, 4R*, 5R*, 7S*, 11R*)-2, (1R*, 4S*, 5R*, 7S*, 11R*)-2 were compared with experimental NMR. The linear correlation analysis towards the experimental and the calculated 13C NMR and DP4+ probability analysis results were shown in Fig. 6. With a DP4+ probability of approximately 100 %, the relative configuration of compound 2 was assigned as 1R*, 4R*, 5R*, 7S*, 11R*. And the absolute configuration was determined as 1R, 4R, 5R, 7S, 11R by comparing experimental and calculated ECD spectra (see Fig. 7).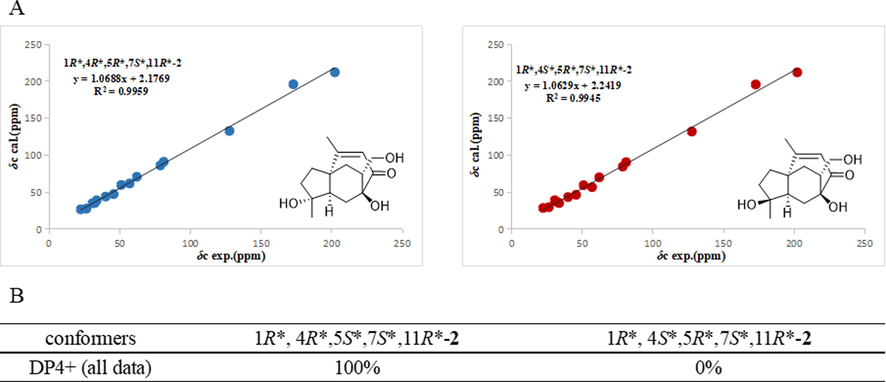
The NMR calculation results of four plausible conformers at the mPW1PW91/6-311+G(d,p) level with IEFPCM in DMSO‑d6 solvent. (A) Linear correlations between the scaled calculated and experimental 13C NMR chemical shifts for compound 2. (B) DP4+ probability of 1H and 13C NMR chemical shifts of 2.
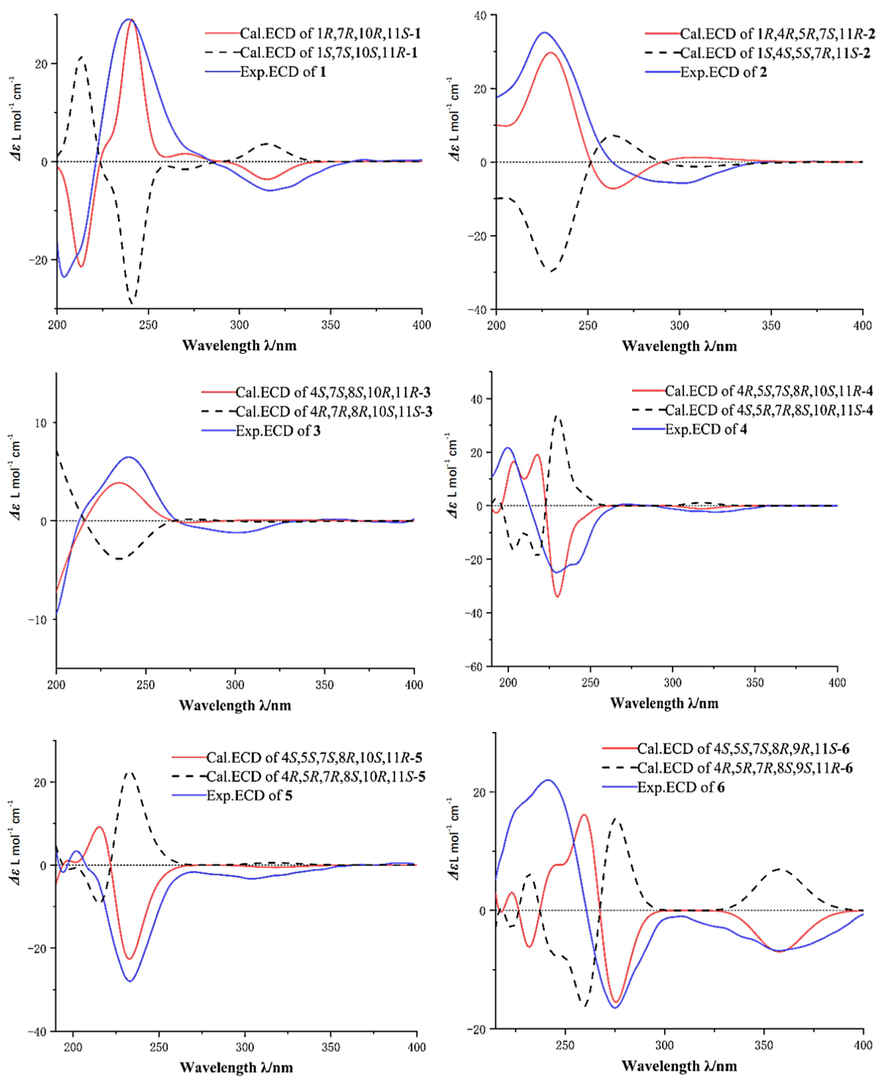
Calculated and experimental ECD spectra of compounds 1–6.
Daphnegenol C (3) was obtained as colorless crystal, and the HRESIMS data (m/z 267.1587 [M+H]+, calcd for 267.1591) gave the molecular formula C15H22O4. Careful inspection of the 1H and 13C NMR spectra revealed that compound 3 was similar to chamaejasmone A (12) (Qiao et al., 2012). The HMBC correlations between H-12 and C-7, C-10, C-11, C-13 supported similarity structure, C-11 was linked to C-10 and C-7. But a hydroxyl in compound 3 and a carbonyl in compound 12, and the location of the double bond was assigned via the HMBC correlations between H-9 and C-1 and between H-6 and C-5. The relative configuration of compound 3 was confirmed by the NOESY correlation of H-15 (δH 1.07)/H-6β (δH 2.28), H-6β (δH 2.28)/H-12 (δH 0.83). The configuration of C-4 and C-8 could not be confirmed by NOESY spectrum, but the single crystal of compound 3 was obtained fortunately. The X-ray diffraction of compound 3 determined the relative and absolute configuration, and the ECD spectrum also supported the absolute configuration as 4S, 7S, 8S, 10R, 11R (see Fig. 7).
Daphnegenol D (4) and daphnegenol E (5) were also yellow syrup and showed the same molecular formulas of C15H22O4 via the HRESIMS (m/z 289.1412 [M+Na]+ and m/z 289.1411 [M+Na]+, calcd for 289.1410) data. The NMR data of compounds 4 and 5 were similar to those of the known compound 12. Detailed analysis of the 1H-1H COSY, HSQC, and HMBC spectroscopic data supported the proposed planar structures for 4 and 5 as shown. Based on the NOESY spectra data and structure model, the NOESY correlation revealed the relative configurations of compound 4 and compound 5 were different at C-4. The relative configuration of C-8 was determined by NMR calculation. The relative configurations of 4 and 5 were determined as 4R*, 5S*, 7S*, 8S*, 10S*, 11R* and 4S*, 5S*, 7S*, 8S*, 10S*, 11R*. And the absolute configurations of compound 4 was determined as 4R, 5S, 7S, 8S, 10S, 11R and compound 5 was determined as 4S, 5S, 7S, 8S, 10S, 11R via ECD comparation.
Daphnegenol F (6) was purified as yellow syrup. It HRESIMS gave a pseudomolecular ion peak at m/z 305.1346[M+Na]+, (calcd for 305.1359) corresponding to the molecular formula of C15H22O5. The structure was similar to the known compound genkwanoid H (Feng et al., 2014) The existence of C-9-O-C-12 moiety was determined by the strong HMBC correlation of H-12/C-9. The NOESY correlation and NMR calculation showed that the relative configuration was identified as 4S*, 5S*, 7S*, 8R*, 9R*, 11S*. The absolute configuration of 6 was established as 4S, 5S, 7S, 8R, 9R, 11S based on ECD calculation.
The HRESIMS of daphnegenol G (7) showed an [M+Na]+ peak at m/z 291.1555 (calcd for C15H24O4Na, 291.1567), combined with 1H NMR and 13C NMR, the molecular formula was deduced to be C15H24O4 with four degrees of unsaturation. The 1D NMR spectra of 7 exhibited one carbonyl carbon, two olefinic carbons, three methyls, five methylenes, two methines and two quaternary carbons. The HMBC spectrum revealed the correlations of three exchangeable protons to the corresponding nearby oxygenated methines, OH-10/C-1, C-10, C-9, OH-13/C-11, C-13 and OH-11/C-7, C-11, C-13. There are characteristic HMBC correlations of H-9/C-7, C-1 and H-6/C-8, C-1, C-4 and H-2/C-5 indicated compound 7 was a guaiane-type sesquiterpenoid. The key NOESY correlations from H-2β (δH 2.42) to H-14 (δH 0.61) and from H-2α (δH 2.28) to OH-1 (δH 5.20) suggested the Me-14 and OH-1 were located on the opposite face. Additionally, the relative configuration of 7 was defined as 1R*, 7R*, 10S*, 11S* via NMR calculation. The absolute configuration of 7 was established as 1R, 7R, 10S, 11S based on ECD calculation (see Fig. 8).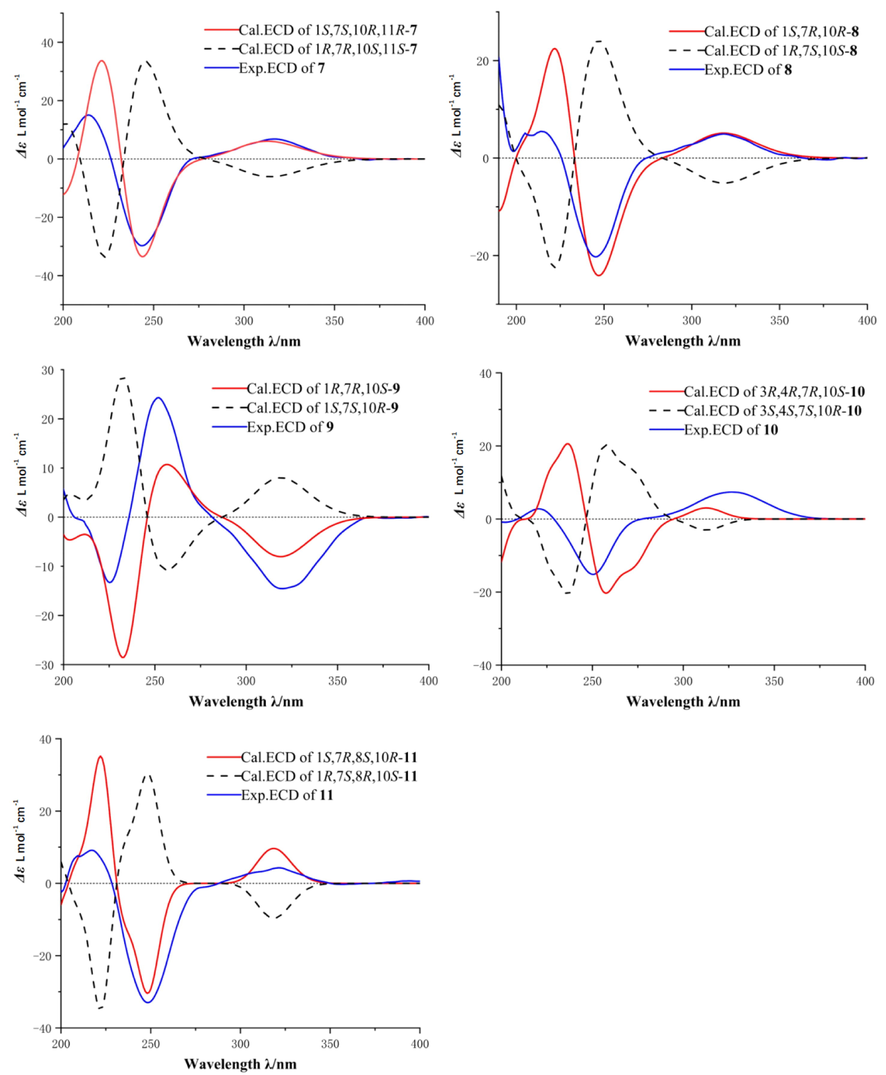
Calculated and experimental ECD spectra of compounds 7–11.
Daphnegenol H (8) and daphnegenol I (9) were obtained as yellow syrup and have the same molecular formula C15H22O4. Compared with the 13C NMR data of 7, compounds 8 and 9 have two more olefinic carbons and lose two aliphatic carbons, and have one more degree of unsaturation. The gross structures of compounds 8 and 9 bearing a terminal double bond at C-11 were elucidated by the set of the HMBC correlations of H-12 with C-7, C-11, C-13. The relative configurations of compounds 8 and 9 were assigned as 1S*, 7R*, 10R* and 1R*, 7R*, 10S*, by the NOESY correlations and DP4+ analyze of calculated NMR data. The absolute configurations of 8 and 9 were established as 1S, 7R, 10R and 1R, 7R, 10S based on ECD calculation, respectively.
The molecular formula of daphnegenol J (10), was determined as C15H22O4 by the HRESIMS m/z 289.1407 [M+Na]+, calcd for 289.1410. The 1H NMR and 13C NMR data reveled that compound 10 also was a guaiane-type sesquiterpenoid. Comprehensive NMR spectroscopic features suggested that hydroxy groups attached at C-3, C-4 and C-13, double bonds at C-1 (C-5) and C-11 (C-12) and carbonyl at C-2. In the NOESY spectrum, the NOE correlations of H-15 (δH 1.07)/H-6α (δH 2.61) and H-14 (δH 0.97)/H-6β (δH 2.42) showed Me-15 and Me-14 located on the opposite face. To ascertain the proposed relative configuration, DFT NMR calculations were systematically performed on the four possible diastereoisomers which could assign the relative configuration of compound 10 as 3R*, 4R*, 7R*, 10S*. Further ECD calculations showed the absolute configuration of 10 was established as 3R, 4R, 7R, 10S.
Daphnegenol K (11) gave the molecular formula as C15H22O4, which could be deduced from the HRESIMS (m/z 289.1415 [M+Na]+, calcd for 289.1410) data. Compared the 13C NMR data with compound 8 and analysis of HMBC data revealed that compound 11 is the hydroxy substitution of compound 8. The stereochemistry of C-8 was established to be S* on the basis of NMR calculation and DP4+ analyze. Finally, the ECD calculation allowed the assignment of the absolute configuration as 1S, 7R, 8S, 10R for 11.
The stereo configuration and NMR data of compounds 8–11 showed a special phenomenon. When H-7 is subject to the spatial effect of Me-14 or Me-15, the chemical shift of H-7 would below 2.50 ppm. Compounds 8 and 9 have same gross structure, in compound 8, the chemical shift of H-7 at 2.84 ppm in DMSO‑d6, but at 2.00 ppm in compound 9. The disparity of it was presumed to the influence of the different orientation of Me-14. The spatial effect of Me-14 leads to the chemical shift of H-7 move to the lower region. After determining the stereo configuration of compound 10, the result showed that Me-15 and H-7 at same orientation, therefore, the chemical shift of H-7 at 1.84 ppm. Compound 11 also conform to the above phenomenon.
Compound 12 was obtained as colorless crystal, compared the 1H and 13C NMR data with literature, it was identified as chamaejasmone A (Qiao et al, 2012). In addition, the configuration of 12 was elucidated by the X-ray diffraction analysis for the first time (see Fig. 5).
Daphnegenol A (1) and daphnegenol B (2) exhibited rare frameworks with 5/6/7 tricyclic system and the plausible biosynthetic pathway was proposed (Cheng et al., 2021). As the precursor of guaiane-type sesquiterpenoids, farnesyl diphosphate (FPP) was substituted by multiple hydroxyls via oxidation reactions to yield oxidized guaiane-type sesquiterpenoids (such as daphneaine E and G), which have been isolated from the root of Daphne genkwa in the previous research (Tan et al., 2019; Guo et al., 2020). Daphneaine G undergo oxidation and dehydration reactions to produce a new six membered oxyheterocyclic ring in 1. Three additional hydroxyls were formed in guaiene by multiple oxidation reactions, and then Prototropy and cyclization reactions made the position of double bond changed and terminal double bond attacked the carbocation to form the additional seven membered ring in 2 (Cane, 1999; Dong et al., 2017) (see Fig. 9).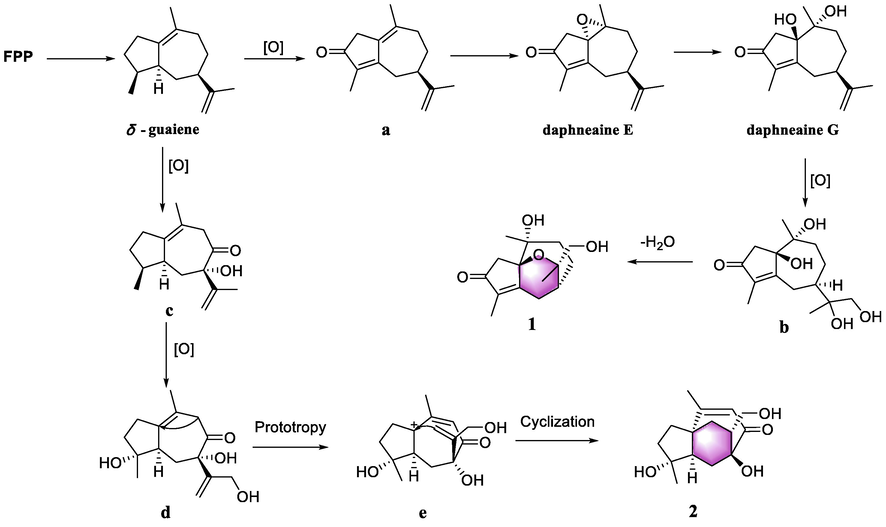
Plausible pathway for the biogenesis of compounds 1 and 2.
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder of brain, and characterized by many pathophysiological indications (Kaur and Bansal, 2022; Guller, 2021). Cholinergic depletion is the major cause and pathological indication of AD (Singh et al., 2020). Modern pharmacological studies have shown that guaiane-type sesquiterpenoids displayed acetylcholineterase (AchE) inhibition activity, which indicated the potential in the treatment of mild to moderate Alzheimer’s disease. The inhibition effect against AchE of all compounds was valued at 100 μM and 10 μM (Ye et al., 2021). These compounds exhibited AchE inhibition activity ranging from 11.9 % to 60.7 %. Compounds 6 and 11 exhibited equivalent activity as positive control donepezil at 100 μM Fig. 10.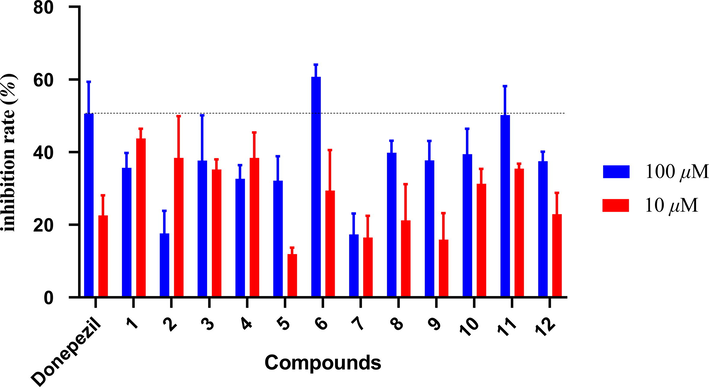
Inhibitory effects of compounds 1–11 against acetylcholinesterase.
In order to further examine the interaction between compounds 6, 11 and AchE, molecular docking studies were carried out to identify the location of the binding sites. Compound 6 and 11 have better bonding in AchE with their docking scores (−7.92 and −7.29, respectively) displayed an approximately equal effect to donepezil (docking score: −8.27), which was consistent with the results of the above inhibition assay. In addition, the in silico experiment results showed that compound 6 bind through hydrogen bond with ASP72 (Choudhary et al., 2005) and Ser 122 (Xu et al., 2020), while compound 11 bind with TYR130 (Sonmez et al., 2017) and HID440 (Bolognesi et al., 2004). HID is a kind of protonated histidine. According to previous studies, ASP72 is the peripheral anionic of AchE, and HID440 is the catalytic triad of AchE (see Fig. 11).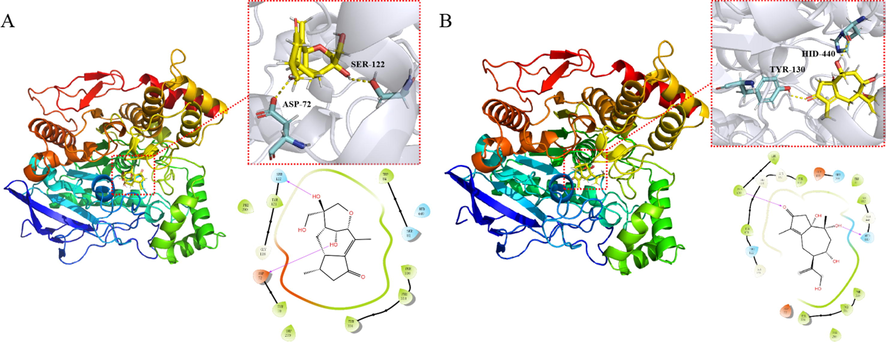
A) Molecular docking of compound 6 with acetylcholineterase. B) Molecular docking of compound 11 with acetylcholineterase.
According previous studies reported, the anti-neuroinflammatory activity also is involved in neurodegenerative diseases such as Alzheimer’s disease (Yang et al., 2021; Elmann et al., 2010). LPS-induced NO overproduction has been suggested to be an important cause in inflammatory effect. All compounds were evaluated the anti-neuroinflammatory activity by measuring the NO concentration. All compounds except compound 3 exhibited moderate anti-neuroinflammatory activity at 10 μM (see Fig. 12).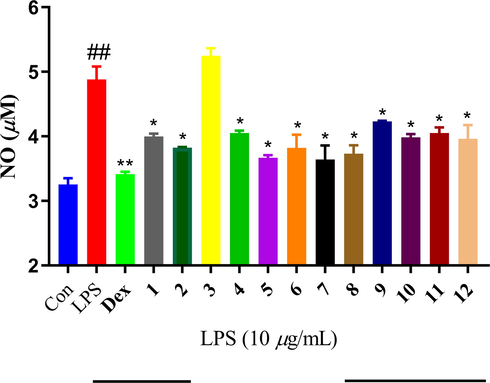
The anti-neuroinflammatory activity of compounds 1–12 with positive control dexamethasone at 10 μM.
4 Conclusion
Based on the molecular networking, eleven undescribed guaiane-type sesquiterpenoids daphnegenol A-K were isolated from the root of D. genkwa. These compounds were identified by comprehensive spectroscopic methods, quantum chemical calculation, and X-ray single crystal diffraction. Daphnegenol A (1) was identified as a guaiane type sesquiterpenoid with a six membered oxyheterocyclic ring and daphnegenol B (2) possessed a rare 5/6/7 tricycles skeleton. All compounds were tested the AchE inhibition and anti-neuroinflammatory activities. Compounds 6 and 11 exhibited equivalent acetylcholineterase inhibition activity as positive control donepezil.
Acknowledgements
This work was financially supported by National Nature Science Foundation of China (82073736, 81872766), Liao Ning Revitalization Talents Program (XLYC2002066), Key Research and Development Project of Liaoning Province (2020JH2/1030058), Science and Technology Planning Project of Liaoning Province (2021JH1/10400049) and Career Development Support Plan for Young and Middle-Aged Teachers in Shenyang Pharmaceutical University (ZQN2018006/ZQN2021011).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- A new sesquiterpenoid from Scaphium macropodum (Miq.) Beumee. Nat. Prod. Res.. 2014;28:597-605.
- [CrossRef] [Google Scholar]
- Chemical studies on the constituents of the Thymelaeaceous plants. IV. Structure of a new spiro biflavonoid, genkwanol A, from the root of Daphne genkwa Sieb. et Zucc. Yakugaku Zasshi. 1987;107:525-529.
- [CrossRef] [Google Scholar]
- Meroterpenoids and sesquiterpene dimers from Sarcandra glabra with anti-neuroinflammatory activity. Ind. Crop. Prod.. 2022;183:114983.
- [CrossRef] [Google Scholar]
- Design, synthesis, and biological evaluation of conformationally restricted rivastigmine analogues. J. Med. Chem.. 2004;47:5945-5952.
- [CrossRef] [Google Scholar]
- Sesquiterpene biosynthesis: Cyclization mechanisms. Compr. Nat. Prod. Chem.. 1999;2:155-200.
- [CrossRef] [Google Scholar]
- Guadiana-type sesquiterpenoids from the roots of Stellera chamaejasme L. and their neuroprotective activities. Phytochemistry. 2021;183:112628.
- [CrossRef] [Google Scholar]
- Juliflorine: A potent natural peripheral anionic-site-binding inhibitor of acetylcholinesterase with calcium-channel blocking potential, a leading candidate for Alzheimer’s disease therapy. Biochem. Bioph. Res. Co.. 2005;332:1171-1179.
- [CrossRef] [Google Scholar]
- Sesqui- and diterpenoids from the radix of Curcuma aromatica. J. Nat. Prod.. 2017;80:3093-3102.
- [CrossRef] [Google Scholar]
- Anti-neuroinflammatory effects of geranium syrup in microglial cells. J. Funct. Foods. 2010;2:17-22.
- [CrossRef] [Google Scholar]
- New sesquiterpenoids from the rhizomes of Acorus tatarinowii. RSC Adv.. 2014;4:42071-42077.
- [CrossRef] [Google Scholar]
- Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Petersson, G.A., Nakatsuji, H., Li, X., Caricato, M., Marenich, A.V., Bloino, J., Janesko, B.G., Gomperts, R., Mennucci, B., Hratchian, H.P., Ortiz, J.V., Izmaylov, A.F., Sonnenberg, J.L., Williams-Young, D., Ding, F., Lipparini, F., Egidi, F., Goings, J., Peng, B., Petrone, A., Henderson, T., Ranasinghe, D., Zakrzewski, V.G., Gao, J., Rega, N., Zheng, G., Liang, W., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Throssell, K., Montgomery, J.A., Jr., Peralta, J.E., Ogliaro, F., Bearpark, M.J., Heyd, J.J., Brothers, E.N., Kudin, K.N., Staroverov, V.N., Keith, T.A., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A.P., Burant, J.C., Iyengar, S.S., Tomasi, J., Cossi, M., Millam, J.M., Klene, M., Adamo, C., Cammi, R., Ochterski, J.W., Martin, R.L., Morokuma, K., Farkas, O., Foresman, J.B., Fox, D.J., 2016. Gaussian, Inc., Wallingford CT.
- Resorcinol derivatives as human acetylcholinesterase inhibitor: an in vitro and in silico study. Int. J. Chem. Technol.. 2021;5:156-161.
- [CrossRef] [Google Scholar]
- Epigenetic manipulation to trigger production of guaiane-type sesquiterpenes from a marine-derived Spiromastix sp. Fungus with antineuroinflammatory effects. J. Nat. Prod.. 2021;84:1993-2003.
- [CrossRef] [Google Scholar]
- Sesquiterpenoids from the roots of Daphne genkwa Siebold et Zucc. with potential anti-inflammatory activity. Phytochemistry. 2020;174:112348.
- [CrossRef] [Google Scholar]
- Neuroprotective compounds from the resinous heartwood of Aquilaria sinensis. Phytochemistry. 2021;181:112554.
- [CrossRef] [Google Scholar]
- Genkwadane F-I, new daphnane-type diterpenes from the flower buds of Daphne genkwa Sieb.et Zucc. exhibit anti-tumor activities via inducing apoptosis. Chin. J. Chem.. 2020;38:1600-1606.
- [CrossRef] [Google Scholar]
- Daphnane-type diterpenes from genus Daphne and their anti-tumor activity. Chin. Herb. Med.. 2021;13:145-156.
- [CrossRef] [Google Scholar]
- Daphnauranols A-C, new antifeedant sesquiterpenoids with a 5/6/7 ring system from Daphne aurantiaca. Tetrahedron Lett.. 2014;55:3693-3696.
- [CrossRef] [Google Scholar]
- Design, molecular docking, synthesis and evaluation of xanthoxylin hybrids as dual inhibitors of IL-6 and acetylcholinesterase for Alzheimer’s disease. Bioorg. Chem.. 2022;121:105670.
- [CrossRef] [Google Scholar]
- Sesquiterpenes from Ulmus davidiana var. japonica with the inhibitory effects on lipopolysaccharide-induced nitric oxide production. Fitoterapia. 2007;78:196-199.
- [CrossRef] [Google Scholar]
- Guaiane-type sesquiterpenoids from Alismatis Rhizoma and their anti-inflammatory activity. Chem. Pharm. Bull.. 2017;65:403-407.
- [CrossRef] [Google Scholar]
- Daphnane-type diterpenes with inhibitory activities against human cancer cell lines from Daphne genkwa. Bioorg. Med. Chem. Lett.. 2013;23:2500-2504.
- [CrossRef] [Google Scholar]
- Shermo: A general code for calculating molecular thermochemistry properties. Comput. Theor. Chem.. 2021;1200:113249.
- [CrossRef] [Google Scholar]
- Advance in biological activities of natural guaiane-type sesquiterpenes. Med. Chem. Res.. 2019;28:1339-1358.
- [CrossRef] [Google Scholar]
- Aquilanols A and B, macrocyclic humulene-type sesquiterpenoids from the agarwood of Aquilaria malaccensis. J. Nat. Prod.. 2017;80:3043-3048.
- [CrossRef] [Google Scholar]
- Guided isolation of daphnane-type diterpenes from Daphne genkwa by molecular network strategies. Phytochemistry. 2022;198:113144.
- [CrossRef] [Google Scholar]
- Guaiane sesquiterpenoids from Torilis japonica and their cytotoxic effects on human cancer cell lines. Arch. Pharm. Res.. 2006;29:131-134.
- [CrossRef] [Google Scholar]
- Guaiane-type sesquiterpenoids from Alisma orientalis. Phytochemistry. 2003;63:877-881.
- [CrossRef] [Google Scholar]
- Neolignans and sesquiterpenes from cell cultures of Stellera chamaejasme. Planta Med.. 2012;78:711-719.
- [CrossRef] [Google Scholar]
- Guaiane-Type Sesquiterpenoids from the roots of Daphne genkwa and evaluation of their neuroprotective effects. J. Nat. Prod.. 2019;82:1510-1517.
- [CrossRef] [Google Scholar]
- Age related neurodegenerative Alzheimer's disease: Usage of traditional herbs in therapeutics. Neurosci. Lett.. 2020;717:134679.
- [CrossRef] [Google Scholar]
- Design, synthesis and docking study of novel coumarin ligands as potential selective acetylcholinesterase inhibitors. J. Enzym. Inhib. Med. Chem.. 2017;32:285-297.
- [CrossRef] [Google Scholar]
- Novel antimalarial guaiane-type sesquiterpenoids from Nardostachys chinensis roots. Tetrahedron Lett.. 1998;39:1361-1364.
- [CrossRef] [Google Scholar]
- Sesquiterpenoids and mycotoxin swainsonine from the locoweed endophytic fungus Alternaria oxytropis. Phytochemistry. 2019;164:154-161.
- [CrossRef] [Google Scholar]
- Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol.. 2016;34:828-837.
- [CrossRef] [Google Scholar]
- Assignment of the stereostructures of sesquiterpenoids from the roots of Daphne genkwa via quantum chemical calculations. Fitoterapia. 2019;138:104352.
- [CrossRef] [Google Scholar]
- Discovery of guaiane-type sesquiterpenoids from the roots of Daphne genkwa with neuroprotective effects. Bioorg. Chem.. 2020;95:103545.
- [CrossRef] [Google Scholar]
- A new flavanol from the roots of Daphne genkwa. J. Asian Nat. Prod. Res.. 2019;21:1215-1220.
- [CrossRef] [Google Scholar]
- Two new guaiane-type sesquiterpenes from Curcuma kwangsiensis and their inhibitory activity of nitric oxide production in lipopolysaccharidestimulated macrophages. Nat. Prod. Res.. 2018;32:2670-2675.
- [CrossRef] [Google Scholar]
- MS/MS-based molecular networking accelerated discovery of germacrane-type sesquiterpene lactones from Elephantopus scaber L. Phytochemistry. 2022;198:113136.
- [CrossRef] [Google Scholar]
- Chemical structures and anti-tyrosinase activity of the constituents from Elephantopus scaber L. Fitoterapia. 2022;162:105259.
- [CrossRef] [Google Scholar]
- Cadinane- and drimane-type sesquiterpenoids produced by Paecilomyces sp. TE-540, an endophyte from Nicotiana tabacum L., are acetylcholinesterase inhibitors. Bioorg. Chem.. 2020;104:104252.
- [CrossRef] [Google Scholar]
- Shenqi Xingnao Granules ameliorates cognitive impairments and Alzheimer’s disease-like pathologies in APP/PS1 mouse model. Chin. Herb. Med.. 2021;12:421-429.
- [CrossRef] [Google Scholar]
- Sesquiterpenoids and tirucallane triterpenoids from the roots of Scorzonera divaricata. Phytochemistry. 2016;124:86-98.
- [CrossRef] [Google Scholar]
- Design, synthesis, and in vitro evaluation of 4-aminoalkyl-1(2H)-phthalazinones as potential multifunctional anti-Alzheimer’s disease agents. Bioorg. Chem.. 2021;111:104895.
- [CrossRef] [Google Scholar]
- iNOS inhibitory activity of sesquiterpenoids and a monoterpenoid from the rhizomes of Curcuma wenyujin. J. Nat. Prod.. 2014;77:2161-2169.
- [CrossRef] [Google Scholar]
- Acetylcholinesterase inhibitors and compounds promoting SIRT1 expression from Curcuma xanthorrhiza. Phytochem. Lett.. 2015;12:215-219.
- [CrossRef] [Google Scholar]
- Three unusual sesquiterpenes with distinctive ring skeletons from Daphne penicillata uncovered by molecular networking strategies. J. Org. Chem.. 2021;86:15298-15306.
- [CrossRef] [Google Scholar]
- Antitumor activity of daphnodorins from Daphne genkwa roots. Int. Immunopharmacol.. 2007;7:128-134.
- [CrossRef] [Google Scholar]
- Isolation and identification of a new dicoumarin from the roots of Daphne genkwa. Acta Pharm. Sin.. 2004;39:990-992.
- [CrossRef] [Google Scholar]
- Absolute configurations and bioactivities of guaiane-type sesquiterpenoids isolated from Pogostemon cablin. J. Nat. Prod.. 2018;81:1919-1927.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.104202.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







