Translate this page into:
Green synthesis and characterization of new carbothioamide complexes; cyclic voltammetry and DNA/methyl green assay supported by silico ways versus DNA-polymerase
⁎Corresponding author. nmmohamed@uqu.edu.sa (Nashwa M. El-Metwaly)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Three novel complexes with the formulae of [Ni(L)2(H2O)2], [Zn(HL)2(OAc)2] and [UO2(L)2].2H2O were synthesized from simple thiosemicarbazide derivative. The ligand in such bis-form binds either as mononegative bidentate or neutral bindentate mode. The NS donors were the binding centers towards all central atoms inside an octahedral geometry. This geometry was supported by 3A2g→3T1g(P,υ3) and 3A2g→3T1g(F,υ2) transitions that assigned from the bands in UV–Vis spectrum of Ni(II) complex. All structures were optimized under DFT method by using B3LYP/6-31G (d) basis set to estimate valuable quantum data. The charges of N11 (-0.197790) and S14 (-0.067486) donors describe their sufficient nucleophilicity for coordination. Cyclic voltammograms of Ni+2 were studied in presence of the ligand to measure the shift in redox peak that affected by the Ni2+-ligand covalency. The elevated stability constants (βML) and Gibbs free energies (ΔG) suggest the successive building to stable complex in solution. The antimicrobial screening showed an excellent inhibition role from the ligand towards the tested bacteria. Moreover, the genotoxicity of the new compounds was assessed using DNA/ methyl green assay. The ligand exhibited the best decay for the green color results from DNA/ methyl green solution. This in vitro investigation was strengthened by silico ways among that the interaction with DNA-polymerase proteins (1bpy, 5szt and 1zqa). A drug with code number MolPort-002–822-252 has high similarity to our ligand in its binding with DNA beside a low Root Mean Square Deviation value (RMSD = 0.266). Further, the ligand interacted perfectly to DG 6(T) residue in 1bpy protein via H-donor bond and the scoring energy value of the docking pose was excellent (S = −6.1512 kcal/mol). The BOILED-Egg indicates the ability of the ligand to absorb in brain barrier. While, there is no ability for such compound to absorb in gastrointestinal tract. Additionally, the ligand doesn't play as a substrate of P-gp protein, which is considered favorable for protecting tissues and organisms.
Keywords
Thiosemicarbazide complexes
DNA/methyl green assay
Cyclic voltammetry
DNA-polymerase
ADME parameters
1 Introduction
Functionally significant biomolecules having S-N donor sites are thiosemicarbazides compounds. These compounds have gotten several importance because of their binding methods, distinctive qualities, biological relevance, and ion-sensing abilities (Casas et al., 2000). In contrast to ligands or metal ions alone, coordinate complexes exhibited high biological activity and features (West et al., 1990). Application areas for carbothioamide-based compounds and their related metal complexes include inorganic chemistry (West et al., 1993, Hosny et al., 2018), material sciences, Langmuir films, electrochemical sensors, as well as dramatically boosting bioactivity (West et al., 1993, Hosny et al., 2018; Liberta and West, 1992, Li et al., 2017). For the treatment of infections and tumors, the industry of drugs needs novel therapeutic medications with broad range of effects with few side effects. Therefore, new sensitive compounds that specifically increase the toxicity of drugs to cancer cells must be developed. On the other hand, the action of small bioactive compounds with DNA can result in genotoxicity, which renders cancer cells inactive. This makes DNA an appealing target for antitumor medicine. Mononuclear complexes were developed by many scientists, and their actions with DNA have been investigated (Abdel-Rhman et al., 2019). DNA proteins and metal ions may attach and interact because DNA has a good electron density whereas metal ions have a low electron density (Abu-Dief et al., 2021). On other side, DNA-polymerase is widely presented enzyme in biological systems that creates complementary DNA strands from template DNA. DNA polymerases are frequently utilized for DNA changes, among that the DNA replication, mutations and other functions (Sumrra et al., 2020), due to their crucial role in preserving genomic fidelity across replication and maintenance. In contrast side, due to their extensive use in a variety of application sectors, especially the biological one as follows, complexes of Schiff base ligands have drawn a lot of attention in recent years. Using a similar ligand extracted with propane hydrazide, complexes were then isolated, the majority of which displayed bioactivity (Alkhamis et al., 2021). The Co(II) and Cd(II) complexes were created employing Schiff base ligand with 1:1 M ratio (Morgan et al., 2017). Investigations into the Ni(II) complex made from thiosemicarbazide's pharmacological effectiveness have largely shown positive results (Leovac et al., 2007). Cobalt and nickel complexes with strong antibacterial activity were made using a thiosemicarbazone derivative with varied molar ratios (Netalkar et al., 2015). The complex has a greater apoptosis efficacy, according to flow cytometric analysis used to measure the cell death brought on by the complexes (Khandar et al., 2019). Copper and nickel thiosemicarbazone complexes were synthesized, studied, and demonstrated distinct antiviral activity (Pelosi et al., 2010). Several metal ion complexes have been created from different thiosemicarbazieds via monobasic or dibasic acids (Miminoshvili, 2009). Hydrazide Ni(II) and Co(II) complexes demonstrated anticancer properties (Antony et al., 2013). Co(II) and Ni(II)-thiosemicarbazone complexes' anticancer properties were also verified (Pelosi, 2010). A green synthesis of copper complex was performed (Alzahrani et al., 2020) using Schiff base derivative. It's important to note that the chemistry could have a big impact on the environment, therefore scientists meticulously examined to make sure that their procedures were as risk-free as feasible. The solvent-free treatment, a green method, was used with ball milling device in several articles (Alzahrani et al., 2020; Alkhamis et al., 2022). The combination of the reagents is firstly validated by the colors created that deviate from the original materials, which happens when they are vigorously crushed in a ball mill without using solvent. After many cleanings with a suitable solvent, the purity of solid products may be assessed (Alkhamis et al., 2021; Alzahrani et al., 2020).
In this study we intended to continue in green synthesis approach for new bivalent complexes that derived from thiosemicarbazide derivative. The new complexes were deliberately analyzed by analytical and spectral tools to characterize their chemical formulae. Quantum based program was implemented to optimize the geometries of all new compounds. Cyclic voltammogram (CV) was studied for the Ni(II) ion in different conditions to examine the degree of stability of the complex formed with the ligand in solution. In vitro, the antimicrobial efficiency of the complexes was investigated as well as their ability to bind with DNA was tested via DNA/methyl green method. This application was supported by utilizing silico methods concerning the most effective compound (the ligand) to investigate the default biological behavior inside living cells.
2 Experimental part
2.1 Reagents
To synthesize thiosemicarbazide derivative as 2-(3,4-dihydronaphthalen-1(2H)-ylidene)-N-ethyl hydrazine-1-carbothioamide (HL), N-ethyl hydrazinecarbothioamide and 3,4-dihydronaphthalen-1(2H)-one were obtained from Sigma & Aldrich. Additionally, on the other side the three complexes were prepared from [Ni(OAc)2].4H2O, [Zn(OAc)2].2H2O and [UO2(OAc)2].2H2O compounds that purchased also from Sigma & Aldrich. Glacial acetic, ethanol (EtOH) and dimethyl sulfoxide (DMSO) were used as they obtained due to their extra-pure nature. The ATCC's agency provided the microbial strains as a holding for biological products and vaccinations. Tris-buffer, MgSO4, and methyl green/DNA have also been purchased from Institute Fermentation of Osaka (IFO).
2.2 Synthesis of the ligand (HL)
The HL ligand was synthesized by mixing N-ethyl hydrazine carbothioamide (0.01 mol, 1.19 g) and 3,4-dihydronaphthalen-1(2H)-one (0.01 mol,1.46 g) in EtOH (50 mL) with addition of glacial acetic acid (three drops) (Alkhamis et al., 2022). The reaction was kept under reflux for 3hr, then the reddish brown precipitate was isolated by filtration and washed by EtOH several times. The dried solid (under CaCl2) was analyzed to estimate its elemental percentages in addition to other analytical data (Table 1). Notice: HL is the abbreviation of the ligand which suggested based on the labile hydrogen existed in thioamide group.
Compound; Empirical Formula (F.W.)
Color
M.p. oC
Yield %
% Found (Calculate)
C
H
M
1
HL; C13H17N3S
247.36Reddish brown
125
83
64.35
(63.12)6.79
(6.90)–
2
[Zn(HL)2(OAc)2]
ZnC30H40N6S2O4; 678.21Yellowish white
190
96
53.22
(53.13)5.97
(5.94)9.70)
(9.64)
3
[Ni(L)2(H2O)2]
NiC26H36N6S2O2; 587.43Dark green
217
98
53.46
(53.16)5.98
(6.18)9.82
(9.99)
4
[UO2(L)2].2H2O
UC26H36N6S2O4; 798.76yellow
>300
96
39.11
(39.09)4.51
(4.54)29.75
(29.79)
2.3 The strategy of green synthesis for the complexes
The weights of reactants applied for green synthesis of complexes were reported in Scheme 1. The molar ratio utilized was 1:1 either from the ligand or the metal ion that mixed together in ball milling technique (Scheme S1) without solvent (Alkhamis et al., 2022). Just a little amount of distilled water was added to facilitate grinding of reactants that continued up to 20 min. The TLC test confirmed that the produced solid complexes were free from reactants after a deliberate washing by distilled water which followed by EtOH. These complexes were technically analyzed to investigate their chemical and structural formulae that appeared by 1metal to 2 ligand.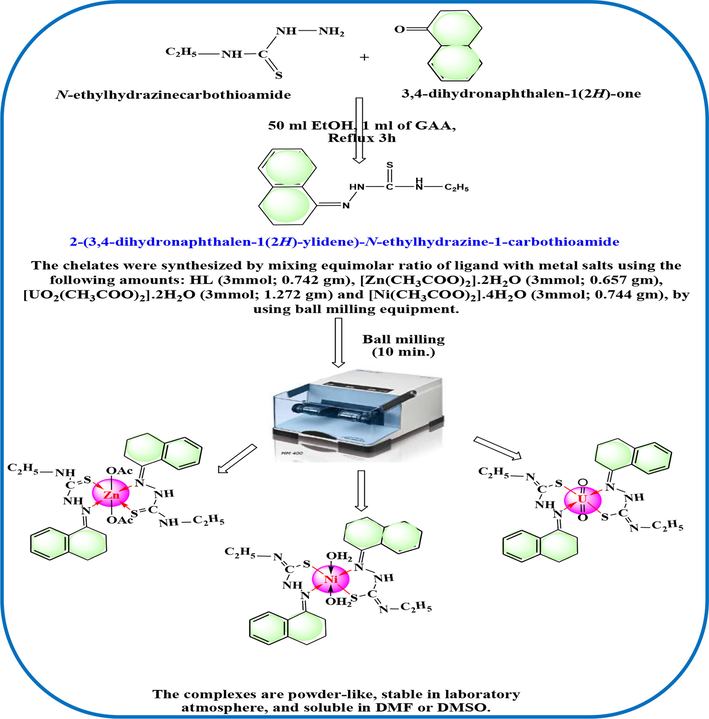
The synthesis strategy y of the ligand (HL) and its metal complexes.
2.4 Equipments of analysis
For simplicity, the majority of the analysis tools were illustrated in the supplementary file as photos, and each tool's specifications were typed beneath its photo (Schemes S2 & S3). The discussion section will also include a separate report on each technique's measuring circumstances. The two ligand' NMR spectra were recorded using Bruker WP (500 MHz) in DMSO‑d6 solvent and TMS as an internal reference. Using JENWAY type 4070 Conductance Bridge, the electrical conductivity of the complexes was assessed for 1 mmol in DMSO solvent. Additionally, through using Johnson Matthey Magnetic Susceptibility Balance, the magnetic moments were calculated by determining the magnetic susceptibility at room temperature. By utilizing a typical procedure for complexometric titration, the Ni(II) and Zn(II) ratios were determined (Vogel, 1989). While the uranium percentage was measured gravimetrically upon complete combustion for weighted sample till reach to U3O8 formula which cooled and then weight.
2.5 Geometry optimization
The Materials Studio software (Modeling and Simulation Solutions for Chemicals and Materials Research and Studio, 2009), the DFT method, and the DNP basis set (Frisch et al., 2009) were all used in conjunction with the DMOL3 tool to improve the structures of most compounds and compute critical parameters. Based on GGA and PBEPBE functions, the best exchange–correlation function was identified (Hehre et al., 1986). Additionally, by using B3LYP/6-31G (d) level on the DFT theory, the MEP map was established. Unfortunately, we couldn't optimize the UO2(II) complex due to selecting of suitable conditions is not easy to reach.
2.6 Cyclic voltammetry
Nickel acetate solution was subjected to this electro-analytical method either in absence or in existence of the coordinating ligand. The cell used is a small beaker which contains 30 mL of 0.L M KCl as a supporting electrolyte. The glassy carbon working electrode (GCWE), Ag/AgCl/KCl (saturated) is the reference electrode, and the platinum wire is an auxiliary electrode used. These electrodes were linked with a potentiostat after being dipped in KCl solution.
2.7 Biological activity
-
Antimicrobial activity screening (MIC)
In accordance with procedure shown in Part A of supporting materials, the disc diffusion method was used to calculate the minimum inhibitory concentration (MIC) for the investigated drugs against various microorganisms (Al-Hazmi et al., 2020). Gram positive (S. aureus & B. subtilis) and Gram negative (E. coli & P. aeuroginosa) bacteria as well as fungi were used to assess the ligand and its associated complexes (C. albicans & A. flavus). As is widely recognized, the term MIC was employed to identify the limiting concentration that was successful in preventing the development of both bacteria and fungus throughout the experiment. The MIC test is the first stage usually employed prior clinical studies in the field of medicines. It involves examining the activity of candidates against target fungi or bacteria.
-
B)
DNA/methyl green binding test
Methyl green reacts reversibly with polymerized DNA; the molecule is stable at neutral pH, whereas the free methyl green decreased. According to the process outlined in Scheme S4 (Burres et al., 1992), the absorption of methyl green had nearly totally vanished after 24 hr in the solution of substitution reaction. Colorimetric technique was used to assess how compounds that can bind to DNA displace methyl green from the molecule. By calculating the IC50 value, which shows the concentration of the compound that causes the absorption to be reduced to half value, the genotoxicity of the currently used compound can be evaluated (Burres et al., 1992).
2.8 Quantum-based silico tests
This part of study aims to confirm the binding efficiency of HL ligand that appeared perfectly invitro assay. Various quantum-based programs were implemented for this virtual study to evaluate and to compare the biological efficiency of HL ligand. The ligand structure was optimized and saved in a suitable format to be used in docking programs (Pharmit link, MOE module & Swiss ADME). The technical details of each methodology were typed in the supporting file (Parts B and C).
3 Results and discussion
3.1 Features of molecular masses
[Ni(L)2(H2O)2], [Zn(HL)2(OAc)2] and [UO2(L)2].2H2O were the formulae of the three complexes derived from thiosemicarbazide derivative (HL)(Table 1). The chemical formulae were suggested based on elemental analysis basically which supported firstly by their incomplete solubility in DMSO or DMF. Such sparingly soluble complexes must have a non-electrolytic nature. The metal (1 mol) to ligand (2 mol) ratio, 1:2, was suggested for all complexes. These bis- ligands were coordinated to complete the stable formulae beside the other secondary ligand either the acetate group or water molecules. A weighted sample (0.2 g) of each complex was kept in oven (at 70 °C) for 1 h and then cooled in dry desiccators under CaCl2. The samples weight is kept constant except the UO2(II) complex (0.191 g). This reduction in the complex weight was calculated and assigned to 2H2O molecules that hydrated the coordination sphere of UO2(II) complex. These samples were returned again to the oven (at 120 °C) for extra 1 h and after cooling they weighted. The Ni(II) complex weight (0.188 g) was reduced in accordance with two coordinating water molecules that decomposed during heating. These observations suggested the presence of crystal water molecules with UO2(II) complex whereas these molecules are covalently attached with nickel atom in the coordination sphere of its complex.
Further, the molecular mass of [Zn(HL)2(OAc)2] complex (i. e.) was confirmed from its mass spectrum (Fig. 1) that performed over m/z range of 50–800, potential difference of 70 eV and heating rate of 40 °C/min. The molecular ion peak was recorded at m/z = 678.07 (calcd. 678.21) which assigns to M+. The successive fragmentation peaks may be assigned as follow in Scheme 2. The isotopes of zinc were recorded at m/z = 66 and 67.![The mass spectrum of [Zn(HL)2(OAc)2] complex.](/content/184/2023/16/7/img/10.1016_j.arabjc.2023.104807-fig2.png)
The mass spectrum of [Zn(HL)2(OAc)2] complex.
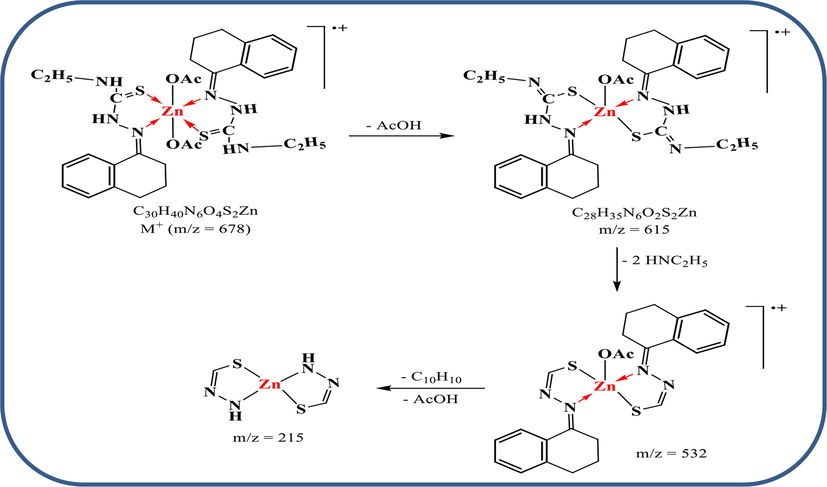
Suggested fragmentation for the mass spectrum of Zn(II) complex.
3.2 Analysis of IR and NMR spectra
Four absorption bands at 3242, 3309, 1049, and 1624 cm−1 in the IR spectrum of HL were assigned to υ(NH)1, υ(NH)2, ʋ(C⚌S), υ(C⚌N) vibrations, respectively (Abdel-Rhman et al., 2019; Abu-Dief et al., 2021; El-Asmy et al., 2015, Hosny et al., 2019). (Table 2, Figure S1). The profile of nuclear magnetic resonance (1H NMR, DMSO‑d6) (Figure S2), showed two signals at 9.80 and 10.91 ppm that were attributed to the protons of (NH)1 and (NH)2. Additionally, there are two signals associated to protons in the ethyl group at 2.52 ppm and 3.47 ppm. Furthermore, several signals in the range of 7.13 to 8.34 ppm signify the aromatic protons. Finally, the spectrum's 13C NMR (Figure S3) revealed two signals at 180.06 and 168.35 ppm, respectively, that are indicative to (C⚌S) and (C⚌N). Moreover, aromatic carbons are responsible for the signals from 124.1 to 140.88 ppm range.
Compound
ʋ(NH)1
ʋ(NH)2
ʋ(C = N)
ʋ(C = S)
ʋ(C = N)*
ʋ(C-S)
ʋ(M−N)
ʋ(M−S)
1
3242
3309
1624
1049
–
–
–
–
2
3333
3377
1596
1011
1631
624
485
551
3
–
3348
1603
–
1637
613
457
502
4
–
3301
1600
–
1620
610
472
525
The IR spectra of [Ni(L)2(H2O)2], and [UO2(L)2].2H2O complexes (Figures S1) revealed a red shift of υ(C⚌N) vibration and disappearance of υ(C⚌S) and υ(NH)1with the instantaneous appearance of υ(C–S) bands at 610–620 and 1615–1625 cm−1 regions (Nakamoto, 1970). This feature suggests a mononegative bidentate mode for each coordinating ligand via the donors of azomethine (C⚌N), and deprotonated thiol (⚌C—S-). The vibrations of new M−L bonds that recorded (Table 2) were assigned to υ(M—N) and υ(M—S) bands (Nakamoto, 1970). Also, ρr(H2O) and ρw(H2O) bands in the Ni(II) complex were appeared at 824 and 529 cm−1, respectively. While, the broadness centered at ≈ 3300 cm−1in the spectrum of UO2(II) complex supports the presence of hydrated water molecules which already suggested (Qurban et al., 2022).
Regarding the UO2(II) complex, the dioxouranium ion exhibited two vibrations, υ3 & υ1, respectively at 906 and 868 cm−1. The force constant (F) of U⚌O bond could being estimated based on McGlynn equation (McGlynn et al., 1961) as follow; (υ3)2 = (1307)2 (FU−O)/14.103. The estimated FU−O value (6.776 mdaynÅ−1) was used to calculate the U-O bond length (RU−O = 1.740 Å) from this equation; RU−O = 1.08 (FU−O)−1/3 + 1.17. The calculated values are within the normal range of uranyl complexes (Qurban et al., 2022; McGlynn et al., 1961).
The IR spectrum of [Zn(HL)2(OAc)2] complex (Figures S1) revealed a red shift of υ(C⚌N), and υ(C⚌S) vibrations which detects the neutral bidentate mode of bonding for each ligand. Azomethine nitrogen and thione sulfur were the coordinating atoms towards the zinc atom. This proposition was confirmed by the appearance of υ(Zn–N) and υ(Zn–S) bands at 485 and 551 cm−1, respectively. Additionally, the vibrations of acetate group were appeared by a difference (Δν > 180 cm−1) refers to its mono-dentate binding nature (Nakamoto, 1970).
On the other hand the NMR spectra of Zn(II) complex were performed in DMSO‑d6 (Figures S2 & S3) to elucidate the suggested structure of the complex. The 1H NMR spectrum showed two signals at δ 9.89, &10.96 ppm which assignable to (NH)1, and (NH)2 protons, respectively. The chemical shifts are almost the same as those of the free ligand, confirming that these groups are not involved in coordination with zinc ion. Moreover, the 13C NMR spectrum of the complex indicated the participation of both (C⚌S), and (C⚌N) in the coordination because they were shifted to down field.
3.3 UV–Vis spectral analysis
Electronic transitions within metal ion complexes are a good indicator for several characteristics based on the field of coordinating ligand. The transitions of intra-ligand and charge transfer are significantly affected after complexation. While, the ligand field transitions within the splitting of d-orbitals are the major indicator for the geometry of paramagnetic complexes (Lever, 1968). So, the UV–Vis spectrum of green Ni(II) complex was done for its solution in DMSO solvent along 200–1000 nm range (Al-Qahtani et al., 2021). The bands recorded at 440 and 735 nm may be assigned to 3A2g→3T1g(P,υ3) and 3A2g→3T1g(F,υ2) transitions, respectively. These transitions are particular to the octahedral geometry of d8 systems. The ligand field parameters as; crystal field splitting energy (Dq = 903), Racah parameter (B = 813) and nephelauetic ratio (β = 0.78) were calculated and found close to the known values of octahedral geometry (Lever, 1968; Al-Qahtani et al., 2021). The nephelauetic ratio reflects the high polarity of new covalent bonds formed between the metal and the ligand. Moreover, the magnetic moment value (μeff = 3.33 BM) is closer to that reported for octahedral form (Cotton et al., 2003).
3.4 Analysis of scan electron microscope (SEM) and EDX patterns
The use of accelerated beam of electrons that focused on solid samples enables to investigate the surface topography of the sample. The SEM images were obtained and displayed for investigation (Figure S4). The image of Zn(II) complex showed a uniform surface of well-spherical particulates that appeared with tiny size in micrometer range. While, the images of Ni(II) and UO2(II) complexes exhibited a non-uniform ice-rocky shape that aggregates various spherical particles (Alkhamis et al., 2021).
The complex's Ni(II), Zn(II), or U(VI) element singlet-set of signals (Figure S5) from the EDX analysis indicate the coordination mechanism properly. For the inner layers to be penetrated in this study, accelerating x-ray photons are necessary (K- or L-shell). These photons stimulate the electrons, which subsequently produce positive gaps that readily draw other electrons from higher levels (L- or M−shell). This relaxation is coupled with radiation emitted that draw peaks of K-series (Ka1, Ka2, or Kb) or L-series. Depending on each element's distinct electronic configuration, these peaks distinguish them; the L-series being the most essential (Alkhamis et al., 2021). The wt/At% was also calculated in relation to the indicator element (heavier element as the metal).
3.5 Geometry optimizing
The geometry of HL ligand and its complexes (Figure S6) except the UO2(II) one was optimized under DFT method. Whereas, 3D-maps (Fig. 2& S7) were demonstrated by using B3LYP/6-31G (d) basis set. These maps as well as the estimated computational parameters (Table S1) offer a well information about the coordination process fully.
-
General features
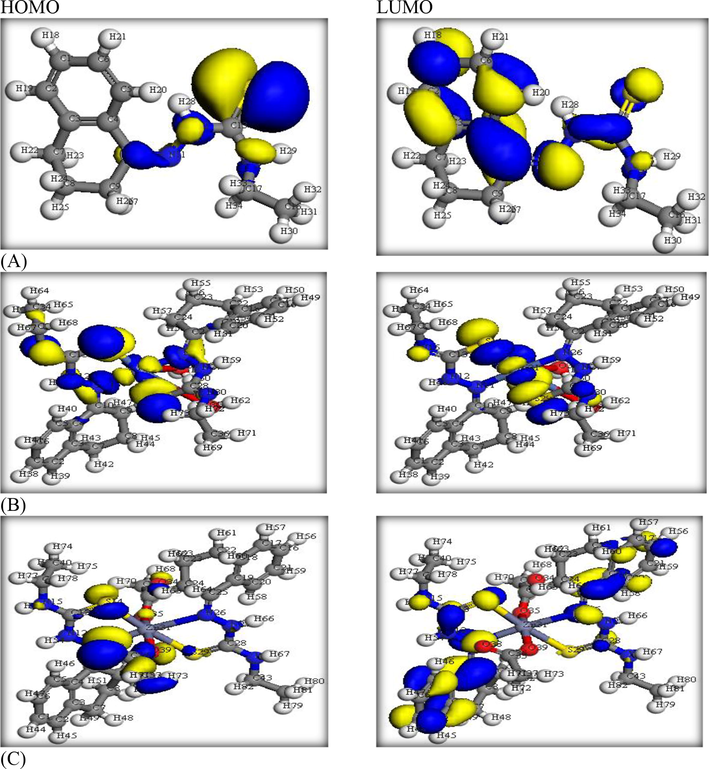
- HOMO & LUMO levels of (A)HL ligand, (B)Ni(II) and (C)Zn(II) complexes.
The ligand geometry of minimum energy content (Figure S6) exhibited a suitable position of C(10) = N(11) and C(13) = S(14) groups that able to coordinate without bond strain. The charges of N11 (-0.197790) and S14 (-0.067486) donors describe their sufficient nucleophilicity for coordination with the metal ions. The bond lengths of C10 = N11 (1.28182 Å) and C13 = S14 (1.58265 Å) groups are significantly lengthen after the coordination of their donors (N & S) towards the metal ions (Alaysuy et al., 2022, Hosny et al., 2023). Further, the angles of S(14)-C(13)-N(12), N(12)-N(11)-C(10) and C(17)-N(15)-C(13) were 122.26°, 121.92° and 122.52°, respectively. These angles reflect the regularity of sp2 hybridized-carbons in HL structure. While such angles were slightly shifted in the two complexes due to some normal changes happened with coordination (Figure S6).
-
B)
3D- models of frontier orbitals and MEP
The HOMO and LUMO patterns were demonstrated (Fig. 2) to evaluate the distribution of electron-cloud on definite functional groups. As seen, the HOMO level in HL ligand was concentrated on the functional groups that contained the donor atoms mainly, while the LUMO level was dispersed over the whole molecule (Alatawi et al., 2022). The feature of the two orbitals did not change strongly in the two complexes while they are seeming focused around the nickel center more than the zinc one. The energies of the two orbitals were computed (Table S1) and the gap between them (ΔE = ELUMO- EHOMO) are significantly lower in the two complexes as expected.
The maps of electrostatic potential (Figure S7) were demonstrated to differentiate the nucleophilic, electrophilic, and zero-potential zones. Such zones were chromatically characterized by red, blue and green colors. As noticed in HL map, the nucleophilic zone (red) seems focused on N(11) and S(14) donors which supporting their priority in coordination among the other donors. This nucleophilicity was reduced in the two complexes depending on the coordination of these donor atoms. Further, the zero-potential zone is normally prevailing on the whole carbon skeleton.
-
C)
Computational parameters
The energy of kinetic, binding, spin electrostatic, polarization and exchange–correlation beside the summation of atomic energy as well as the dipole moments were estimated and tabulated (Table S1). The values of total energy, summation of atomic energy and binding energy of the two complexes are lower than the free ligand indicating the high stability of such complexes. The exchange–correlation energy and electrostatic energy values are also lowered in the complexes to confirm their priority in stability of bonds. The dipole moment of HL ligand and its corresponding complexes are relatively comparable to each other's and their values detect the reduced polarity over the whole molecules. This feature is preferable in biological application due to the ability of such compounds to miscible with cell-lipids and then may contact directly with active sites in biological systems (Abumelha et al., 2020).
3.6 Cyclic voltammetry
This study offers essential parameters that reflect a light on the changes of electrochemical behavior of nickel ion (i. e.) in presence of the ligand. This may help to understand the coordination process within the complexes in solid state as the main aim in this research. Notice: the equations implemented for this study were typed in the supporting material file (part D) to reduce the similarity in the manuscript as possible.
-
Electrochemical behavior of Ni2+ in absence and presence of HL at 288.55 K
The redox behavior of Ni2+in 0.1 M KCl (supporting electrolyte) was studied electrochemically at 288.55 K under glassy carbon electrode (working one), potential window of 1 to −1 V and a scan rate 0.1 V/s (Fig. 3). In absence of HL ligand, the electroactivity of Ni2+ solution was suggested due to appearance of one cathodic and anodic peaks on the voltammogram which attributing to (Ni2+/Ni0) and (Ni0/ Ni2+), respectively. The catiodic potential peak, Epc, arises to −0.4525 V in the forward scan, while the anodic potential peak, Epa, reaches to 0.0425 V in the reverse scan. The increase in all peak currents designates and supports the presence of diffusion process (Almehmadi et al., 2022). Adding HL ligand by various concentrations leads to lower shift of the redox peak due to the covalency between the Ni2+ ions and the ligand (Fig. 3).
-
B)
Stability constant and Gibbs free energies for the Ni2+-HL complex
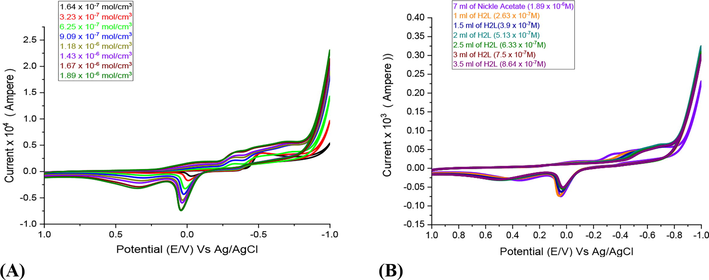
- Influence of different concentrations of Ni(II) in absence of HL (A) and in presence of HL (B) using glassy carbon electrode in 0.1 M KCl at 288.55 K and scan rate 0.1 V.S-1.
Adding HL ligand to Ni2+ ions, the elevated stability constants (βML) and Gibbs free energies (ΔG) suggest the successive building to stable complex in solution as shown in Table 3. To calculate βML and ΔG, we must apply these Eqs. (1)–(3):
[M] x10-3 mol. L-1
[L] × 10-4mol.L-1
(Ep,1/2)M
(Ep,1/2)C
ΔE (V)
J (L/J)
Log βj
ΔG (kJ/mol)
1.84
2.63
−0.205
−0.2141
−0.0091
0.1429
0.3527
−1.949
1.82
3.9
−0.205
−0.2351
−0.0301
0.2142
0.2049
−1.132
1.79
5.13
−0.205
−0.2477
−0.0427
0.2857
0.1943
−1.073
1.77
6.33
−0.205
−0.2553
−0.0503
0.3571
0.2639
−1.458
1.75
7.5
−0.205
−0.2710
−0.0661
0.4285
0.1860
−1.028
1.73
8.64
−0.205
−0.2733
−0.0684
0.5010
0.3376
−1.865
Where (E˚M) is the valid peak potential of the Ni+2 ion at zero ligand concentration, (E˚Ml) is the valid peak potential of the complex after each addition for the ligand amount, (j = [L]/[M]) is the molar ratio, and (CL) is the molar concentration of the ligand. Finally, increasing the ligand concentration the βML and ΔG values are increased.
3.7 Invitro assays
-
Antimicrobial screening
Gram positive (S. aureus & B. subtilis), Gram negative (E. coli & P. aeuroginosa), and fungi (C. albicans & A. flavus) were chosen for this test concerning the HL ligand and its corresponding complexes. The MIC method was applied, and the results were aggregated in Table 4 and graphed in Fig. 4 (Raman et al., 2003). Two references were screened for comparative evaluation, the first is Ampicillin as the antibacterial reference, and the second is Colitrimazole as antifungal reference. The results point to the superiority of the ligand towards all bacteria which appeared more than the reference. Also, the antifungal screening results are also excellent. The antimicrobial activity of Ni(II) complex comes right after the ligand activity. Unfortunately, the behaviour of rest complexes is ineffective. As reported in various researches (Shah et al., 2016), the existence of functional groups containing nitrogen or sulphur atom enables the chemical compound to interact with electron-deficient centres in biological systems. This is noticed by a distinguish way with the free HL ligand, while this feature was reduced with the complexes. This may refer to the coordination of some functional groups with the metal ions which may cause incapability of such groups for other contributions. Fortunately, the activity of Ni(II) complex is highly significant which is due to the toxic role of nickel atom inside living cells.
-
B)
DNA/methyl green test
| Compound | E. coli | P. aeuroginosa | S. aureus | B. subtilis | C. Albicans | A. flavus |
|---|---|---|---|---|---|---|
| Ampicillin | 125 | 187 | 187 | 93 | – | – |
| Colitrimazole | – | – | – | 6 | 4 | |
| 1) HL | 125 | 125 | 125 | 62 | 11 | 6 |
| 2) [Zn(HL)2(OAc)2] | 343 | 337 | 458 | 369 | 73 | 105 |
| 3) [Ni(L)2(H2O)2] | 165 | 161 | 132 | 109 | 21 | 30 |
| 4) [UO2(L)2].2H2O | 338 | 333 | 467 | 366 | 49 | 45 |
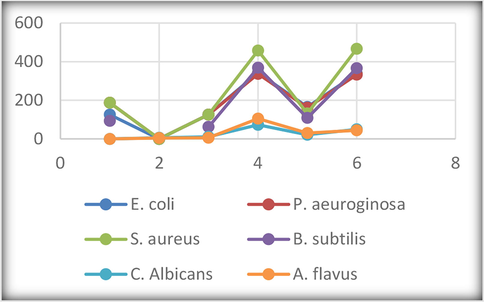
- Antimicrobial activity in terms of MIC (µg/mL).
Although the bioactivity of newly chemicals are investigated from many scientists, understanding the real mechanism happened inside the living cells is still an area of disagreement. But the best way for choosing the pharmaceutical compound is not area of disagreement between them, they concentrated on the binding activity of such compound with the DNA. The affinity of methyl green to stain the DNA via binding at pH = 7 was clearly recorded in a reversible reaction in between (Nagar, 1990). The magnitude of decay in the green color staining the DNA in presence of a tested compound measures the reactivity of such compound in replacing methyl green. This decay was monitored at λ = 630 nm and the concentration of the tested compound causes the absorption reduction to its half value (IC50) was determined for each compound (Table 5). The ligand and [Ni(L)2(H2O)2] complex exhibited significant genotoxicity towards the DNA, particularly the ligand (Ramesh and Maheswaran, 2003). Consequently, controlling the cell-DNA via its combination with the compound is the main target to inhibit cell division, resulting induced apoptosis. When the damage of DNA is becoming serious and cannot easily corrected so the cell will death. This damage based on the deterioration of DNA helix due to intercalation of HL ligand with the grooves or through H-bonding. We know the role of aromatic ring in intercalation with DNA, which considered the first part from the ligand that really interacted (Abdel-Rahman et al., 2021). Prospective research is required to determine whether genetic information can improve therapeutic efficacy and safety by assisting with drug selection and dose. Despite the fact that there still many obstacles to overcome, the integration of pharmacogenetics into clinical practice is anticipated to have a substantial impact on health in the upcoming years. The concentration needed to reduce absorbance of DNA bonded to methyl green to its half value (IC50) was evaluated(mean ± SD, n = 3–5 separate determinations).
DNA-active compound
DNA/methyl green
(IC50, µg/ml)
1
45.3 ± 2.6
2
78.4 ± 1.6
3
57.9 ± 2.3
4
75.8 ± 2.6
3.8 Quantum-based in-silico tests
-
Pharmit server and MOE module
Here we aimed to examine the extent of effectiveness for binding between the ligand and DNA-polymerase to evaluate the degree of conformity with in-vitro results. Three proteins were chosen from PDB as 1bpy, 5szt and 1zqa (Scheme S5) for this docking purpose using either the pharmit server (Daina et al., 2017, Fenton et al., 2002; Maiorov and Crippen, 1994) or MOE module (Ver. 2018). Pharmit server, showed a best interaction for the ligand with the pockets of 1bpy and 5szt proteins (Fig. 5). This simulation was ended by searching on analogous drug in MolPort database, a drug with code number MolPort-002–822-252 (Scheme 3) has extent of similarity with HL ligand in its binding with DNA. The Root Mean Square Deviation value (RMSD = 0.266) of the drug is very small which points to the high similarity of such drug with the tested ligand in their biological effectiveness (Maiorov and Crippen, 1994).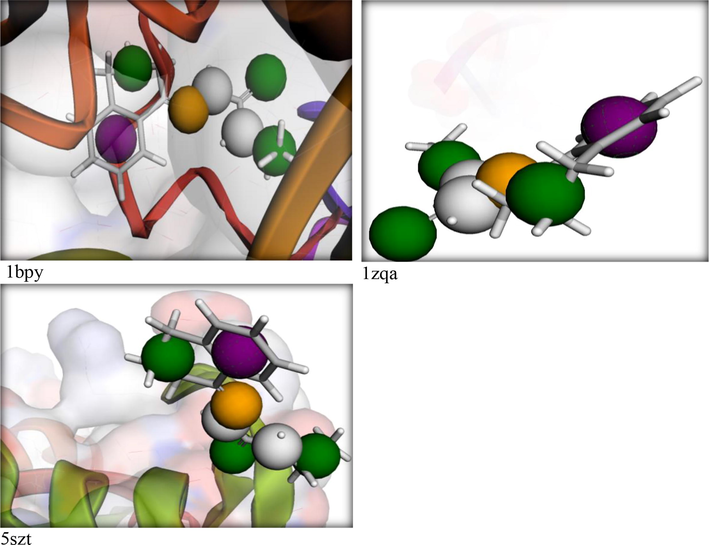
Pharmit interaction for the HL ligand towards DNA-polymerase proteins.
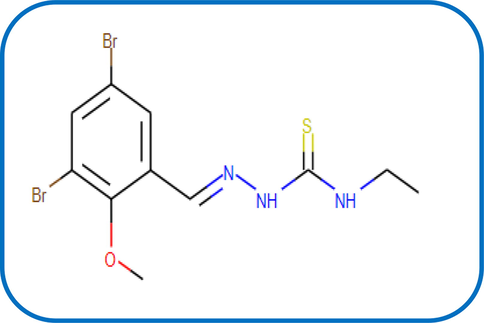
The structure of MolPort-002–822-252.
Such distinguished behavior of the ligand towards DNA-polymerase was confirmed by MOE-docking (Fig. 6). The ligand interacted perfectly to DG 6(T) residue in 1bpy protein via H-donor bond (length 3.3 Å) and the scoring energy value of the docking pose was excellent (S = −6.1512 kcal/mol). While, the allosteric binding with 5szt protein was conducted through various aminio acid residues to produce a pose also with excellent scoring energy (−5.9887 kcal/mol) (Hosny et al., 2020; Katouah et al., 2020).
-
B)
ADME parameters
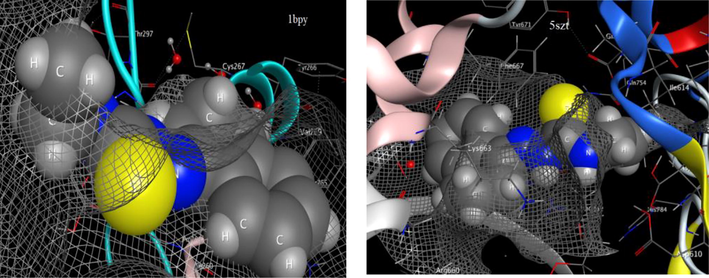
- Effective Interaction of HL with two DNA-polymerase proteins.
The absorption, distribution, metabolism and excretion (ADME) are the parameters targeted to investigate the ligand (smaller size) by using Swiss-link. The bioavailability radar (Fig. 7) which exhibited XLOGP3 (lipophilicity), polarization, saturation limit (sp3 ≥ 0.25), size, flexibility and solubility indexes on six axis reflects the drug-likeness and physicochemical properties of the ligand. The pink region detects the limits of the optimum values of such indices (Alkhamis et al., 2021). The values of XLOGP3 (2.61), saturation (0.38), size (247.3), polarity (TPSA = 68.51), and flexibility (4) are lower enough to judge on distinguish HL drug-like and physicochemical properties. The lipophilic feature of the ligand was also supported by LogPo/w (2.74) and LogS (-3.02) values that indicate the lower solubility of HL in polar solvent as water (Alkhamis et al., 2021).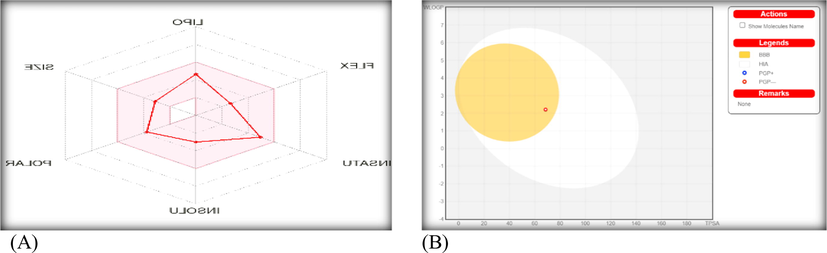
The bioavailability radar (A) and BOILED-Egg (B) for the ligand via Swiss-ADME study.
Additionally, the permeability of skin (Log Kp), P-glycoprotein (P-gp) substrate, gastrointestinal tract absorption (GI), brain barrier absorption (BBB), and cytochromes P450 (CYP) were the indices of ligand pharmacokinetics. The behavior of HL ligand towards P-glycoprotein (P-gp) and cytochrome P450 (CYP) enzymes was investigated based on the importance of such protein (P-gp) or these enzymes. The P-gp could protect the nerve cells from xenobiotics although its excretion in tumor cells will resist the anticancer drug action. On the other side, a group of cytochromes which is called iso-enzymes (CYP1A2, CYP2C19, CYP2C9, CYP2D6 and CYP3A4) is essential for de-accumulation process (via metabolism) of remaining drug inside the cell (Abu-Dief et al., 2021). The HL ligand doesn't play as a substrate of P-gp protein, which is considered favorable for protecting tissues and organisms. Further, the ligand did not inhibit three CYP-enzymes just inhibit CYP1A2 & CYP2C19. So, the drug metabolism process inside the cell did not affected seriously. The log Kp (-5.96 cm/s) that reflects the relation between lipophilicity and size of the compound, appeared with smaller negative value, indicates the high extent of skin permeability for dug.
Furthermore, the BOILED-Egg image (Fig. 7) which point to a relation between WLOGP and TPSA, indicates the ability of the ligand (red point) to absorb in brain barrier as appeared in yellow region (yolk) (Abu-Dief et al., 2021). While, there is no ability for such compound to absorb in gastrointestinal tract (white zone). Also, the red point reflects the non-substrate function of the ligand towards P-gp (PGP-) protein. The interaction pathway of the ligand inside the living cells (Figure S8) was oriented towards cathepsin L protease; leukocyte elastase protease by 26.7 %, P2X purinoceptor 7 (by homology) ligand-gated ion channel by 20.0 %, and isocitrate dehydrogenase [NADP] cytoplasmic enzyme by 13.3 %. Finally, the interaction of the HL ligand with DNA-polymerase cannot be ignored either from in vitro or in silico evaluations. This study may be the out of point for other extensive studies aimed at deepening the biological efficacy of the carbothioamide derivative and enriching the rich history of this family of compounds with biological excellence.
4 Conclusion
Three metal ion complexes were derived from thiosemicarbazide derivative by a molar ratio of 1: 2 for metal to ligand. The chemical formulae of all compounds were elucidated by elemental analysis, IR, NMR, UV–Vis, mass spectrum, SEM and EDX and also supported by quantum calculations. The ligand coordinates via C(10) = N(11) and C(13) = S(14) groups either as neutral form or as monobasic form. The cyclic voltammetry of Ni+2 solution was studied in absence and presence of the ligand to understand the electrochemical changes that happened due to complexation. The high stability constants (βML) and Gibbs free energies (ΔG) suggest the presence of stable complex in solution. In vitro assessments were performed on the new compounds against various microbes as well as methyl green/ DNA-binding efficiency. All the results showed exceeding biological efficiency of the ligand moreover appeared superior the antibacterial reference drug. The genotoxicity of the ligand was confirmed via silico tests towards the interaction with DNA-polymerase proteins (1bpy, 5szt and 1zqa). Also, the BOILED-Egg indicates the probability of ligand absorption in brain barrier and there is no probability for absorption in gastrointestinal tract. Finally, this study may be the outset point for other extensive studies aimed at deepening the biological efficacy of the carbothioamide derivative and enriching the rich history of this family.
Acknowledgement
Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R22), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Synthesis, characterization and in vitro pharmacological evaluation of 6,6'-((1E,1'E)-((4-chloro-1,2-phenylene) bis (azaneyl ylidene)) bis(methaneylylidene)) bis (2-ethoxy phenol and its Zr(IV), V(IV), Zn(II) chelates as promising antiproliferative, antimicrobial, antioxidant drug candidates. Bioorg. Chem.. 2021;114:105106
- [Google Scholar]
- Synthesis, characterization, molecular docking and cytotoxicity studies on N-benzyl-2-isonicotinoylhydrazine-1-carbothioamide and its metal complexes. J. Mol. Struct.. 2019;1196:417-428.
- [Google Scholar]
- Synthesis and intensive characterization for novel Zn (II), Pd (II), Cr (III) and VO (II)-Schiff base complexes; DNA-interaction, DFT, drug-likeness and molecular docking studies. J. Mol. Struct. 2021:130693.
- [Google Scholar]
- Targeting ctDNA binding and elaborated in-vitro assessments concerning novel Schiff base complexes: synthesis, characterization, DFT and detailed in-silico confirmation. J. Mol. Liq.. 2021;322:114977.
- [Google Scholar]
- deliberate-characterization for Ni (II)-Schiff Base complexes: promising in-vitro anticancer feature that matched MOE docking-approach. J. Inorg. Organomet. Poly. Mater.. 2020;30(9):3277-3293.
- [Google Scholar]
- Simulation for the behavior of new Fe (III) and Cr (III)-thiophenyl complexes towards DNA polymerase: synthesis, characterization, eukaryotic DNA and Hartree-Fock computation. Chem. Pap.. 2022;76(6):3919-3935.
- [Google Scholar]
- Elucidating of new hydrazide-based complexes derived from Pd (II), Cu (II) and Cd (II) ions: studies concerning spectral, DFT, Hirshfeld-crystal, biological screening beside Swiss-ADME verification. J. Mol. Struct.. 2022;1259:132748.
- [Google Scholar]
Synthesis, characterization, DFT, cytotoxicity evaluation and molecular docking of a new carbothioamide ligand and its coordination compounds. Results Chem.. 2023;5:100776.- [Google Scholar]
- Green synthesis approach for Fe (III), Cu (II), Zn (II) and Ni (II)‐Schiff base complexes, spectral, conformational, MOE‐docking and biological studies. Appl. Organomet. Chem.. 2020;34(3)
- [Google Scholar]
- Elucidation for coordination features of hydrazide ligand under influence of variable anions in bivalent transition metal salts; green synthesis, biological activity confirmed by in-silico approaches. J. Mol. Struct.. 2021;1238:130410
- [Google Scholar]
- Conductometry of nano-sized zinc sulfate; synthesis and characterization of new hydrazone complexes: conformational and in-vitro assay. J. Mol. Liq.. 2021;340:117167
- [Google Scholar]
- Conductometry of nano-sized zinc sulfate; synthesis and characterization of new hydrazone complexes: conformational and in-vitro assay. J. Mol. Liq.. 2021;340:117167
- [Google Scholar]
- Synthesis and investigation of bivalent thiosemicarbazone complexes: conformational analysis, methyl green DNA binding and in-silico studies. Arab. J. Sci. Eng.. 2022;1–18
- [Google Scholar]
- Solvent free synthesis, characterization, DFT, cyclic voltammetry and biological assay of Cu (II), Hg (II) and UO2 (II)–Schiff base complexes. Arb. J. Chem. 2022;15(2):103586.
- [Google Scholar]
- Green synthesis for new Co (II), Ni (II), Cu (II) and Cd (II) hydrazone-based complexes; characterization, biological activity and electrical conductance of nano-sized copper sulphate. J. Mol. Struct.. 2021;1244:131238
- [Google Scholar]
- Ball milling approach to prepare new Cd (II) and Zn (II) complexes; characterization, crystal packing, cyclic voltammetry and MOE-docking agrees with biological assay. J. Mol. Struct.. 2020;1218:128473
- [Google Scholar]
- Synthesis, spectrochemical characterisation and catalytic activity of transition metal complexes derived from Schiff base modified chitosan. Spectrochim. Acta A. 2013;103:423-430.
- [Google Scholar]
- A colorimetric microassay for the detection of agents that interact with DNA. J Nat Prod. 1992;55:1582-1587.
- [Google Scholar]
- Main group metal complexes of semicarbazones and thiosemicarbazones. A structural review. Coordination Chem. Rev.. 2000;209(1):197-261.
- [Google Scholar]
- Advanced Inorganic Chemistry (6th ed.). John Wiley & Sons Inc; 2003. p. :1-171.
- Sci. Rep.. 2017;7:42717. Macrocyclic ligand design. Structure–function relationships involving the interaction of pyridinyl-containing, mixed oxygen–nitrogen donor macrocycles with cobalt (II), nickel (II), copper (II), zinc (II), cadmium (II), silver (I) and lead (II) J. Chem. Soc. Dalton Trans.. 2002;10:2185-2193.
- [Google Scholar]
- Spectral, thermal, molecularmodeling and biological studies on monoandbinuclear complexes derived from oxalobis(2,3-butanedionehydrazone) Chem. Cent. J.. 2015;9:1-12.
- [Google Scholar]
Synthesis, characterization and cytotoxicity of new 2-isonicotinoyl-N-phenylhydrazine-1-carbothioamide and its metal complexes. Appl. Organomet. Chem.. 2019;33(8):e4998.- [Google Scholar]
- Frisch, M.J., Trucks, G.W., Pople, J.A., 2009. Gaussian 09, Revision B.2, Gaussian, Inc., Pittsburgh, PA.
- Ab Initio Molecular Orbital Theory. New York: Wiley; 1986.
- Synthesis, Spectral, Modeling, Docking and Cytotoxicity Studies on 2-(2-aminobenzoyl)-N-ethylhydrazine-1-carbothioamide and its divalent metal complexes. Appl. Organomet. Chem.. 2020;34(11):e5922.
- [Google Scholar]
- Facile synthesis and deliberate characterization for new hydrazide complexes; cyclic voltammetry, crystal packing, eukaryotic DNA degradation and in-silico studies. J. Mol. Liq.. 2020;320:114380
- [Google Scholar]
- Cadmium(II) complexes of a hydrazone ligand: Synthesis, characterization, DNA binding, cyto- and genotoxicity studies. Polyhedron. 2019;171:237-248.
- [Google Scholar]
- Transition metal complexes with thiosemicarbazide-based ligands. Part LVI: Nickel(II) complex with 1,3-diphenylpyrazole-4-carboxaldehyde thiosemicarbazone and unusually deformed coordination geometry. Polyhedron. 2007;26(14):3783-3792.
- [Google Scholar]
- Inorganic Electronic Spectroscopy. Amsterdam: Elsevier; 1968. p. :1-863.
- Antifungal and antitumor activity of heterocyclic thiosemicarbazones and their metal complexes: current status. Biometals. 1992;5(2):121-126.
- [Google Scholar]
Improved thermal lattice Boltzmann model for simulation of liquid-vapor phase change. Phys. Rev. E. 2017;96:063303- [Google Scholar]
- Significance of root-mean-square deviation in comparing three-dimensional structures of globular proteins. J. Mol. Biol.. 1994;235(2):625-634.
- [Google Scholar]
- Electronic structure, spectra, and magnetic properties of oxycations. III. Ligation effects on the infrared spectrum of the uranyl ion. J. Chem. Phys.. 1961;35(1):105-116.
- [Google Scholar]
- Modeling and Simulation Solutions for Chemicals and Materials Research, Materials Studio (Version 5.0), 2009. Accelrys software Inc., San Diego, USA. Available online at: http://www.accelrys.com.
- Geometrical structures, thermal properties and spectroscopic studies of Schiff base complexes: Correlation between ionic radius of metal complexes and DNA binding. J. Mol. Liq.. 2017;240:752-776.
- [Google Scholar]
- Syntheses, characterization, and microbial activity of some transition metal complexes involving potentially active O and N donor heterocyclic ligands. J Inorg Biochem. 1990;40:349-356.
- [Google Scholar]
- Infrared Spectra of Inorganic and Coordination Compounds. New York: Wiley; 1970. p. :1-338.
- Transition metal complexes of thiosemicarbazone: Synthesis, structures and invitro antimicrobial studies. Polyhedron. 2015;100:215-222.
- [Google Scholar]
- Thiosemicarbazone metal complexes: from structure to activity. Open Crystallogr. J.. 2010;3:16-28.
- [Google Scholar]
- Maria Carla Re, Claudio Casoli, Elisabetta Pilotti, Antiretroviral activity of thiosemicarbazone metal complexes. J. Med. Chem.. 2010;53(24):8765.
- [Google Scholar]
- Tailoring of new Ni(II), Hg(II) and UO2(II)–hydrazide complexes: characterization, studies in-vitro and in-silico as well as the Hartree-Fock modeling. J. Saudi Chem. Soc.. 2022;26(3):101477
- [Google Scholar]
- Synthesis, characterization and electrochemical behaviour of Cu(II), Co(II), Ni(II) and Zn(II) complexes derived from acetylacetone and p-anisidine and their antimicrobial activity. Proc. Indian Acad. Sci.. 2003;115:161-167.
- [Google Scholar]
- Synthesis, spectra, dioxygen affinity and antifungal activity of Ru(III) Schiff base complexes. J. Inorg. Biochem.. 2003;96:457-462.
- [Google Scholar]
- Elaborated studies on nano-sized homo-binuclear Mn (II), Fe (III), Co (II), Ni (II), and Cu (II) complexes derived from N2O2 Schiff base, thermal, molecular modeling, drug-likeness, and spectral. J. Therm. Anal. Calorim.. 2016;123(1):731-743.
- [Google Scholar]
- A review on the efficacy and medicinal applications of metal-based triazole derivatives. J. Coord. Chem.. 2020;73(20–22):2838-2877.
- [Google Scholar]
- Quantitative Inorganic Analysis. London: Longmans; 1989.
- Structural and physical correlations in the biological properties of transition metal heterocyclic thiosemicarbazone and S-alkyldithiocarbazate complexes. Struct. Bonding. 1990;76:1-50.
- [Google Scholar]
- Thiosemicarbazone complexes of copper (II): structural and biological studies. Coordination Chem. Rev.. 1993;123(1–2):49-71.
- [Google Scholar]
Spectral, optical, and cytotoxicity studies on N-(2-isonicotinoylhydrazine-carbonothioyl)benzamide and its metal complexes. Heteroatom Chem.. 2018;29(2):e21415.- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.104807.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







