Translate this page into:
Mechanistic study on DNA mutation of the cytosine methylation reaction at C5 position
⁎Corresponding authors at: Department of Chemistry, University of Jordan, Amman 11942, Jordan (M.H. Almatarneh). Department of Chemistry, Memorial University, St. John’s, NL A1B 3X7, Canada (Y. Zhao). m.almatarneh@ju.edu.jo (Mansour H. Almatarneh), yuming@mun.ca (Yuming Zhao)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
DNA methylation is a crucial epigenetic mark connected to conventionally changing the DNA bases, typically by adding methyl groups into DNA bases. Methylation of cytosine at the C5 position (5-methylcytosine) occurs mostly in the context of cytosine-phosphate-guanine dinucleotides, the methylation of which has important impacts on gene regulation and expression. However, the mechanistic details of this reaction are still debatable concerning the concertedness of the key reaction steps and the roles played by the base that abstracts the proton in the β-elimination and water molecules at the active site. To gain a deeper insight into the formation of 5-mehtylcytosine, an extensive density functional theory (DFT) study was performed with the B3LYP functional in conjunction with different basis sets. Our study has clearly established the mechanistic details of this methylation approach, based on which the roles of conserved active site residues, such as glutamic acid and waters, are well understood. Our results show that the reaction of 5-methylcytosine follows a concerted mechanism in which water molecules are critically involved. Moreover, arginine and alanine give more significant catalytic effects than glutamic acid on the 5-methylcytosine process. Considering the effect of Alanine, Arginine, and one water bridging molecule, the activation energy is 31 kJ mol−1 calculated at B3LYP/6-31G(d) level of theory.
Keywords
Epigenetics
DNA Methylation
DNA Methyltransferases
5-Methyl Cytosine
CpG methylation
Transition state
1 Introduction
Methylation of cytosine at the C5 position is an epigenetic modification linked to many human diseases. In eukaryotes, methylation takes place at positions 5 and 4 of cytosine produces 5-methylcytosine (5mC) and 4-methylcytosine (4mC), respectively, while methylation at position 6 in adenine yields 6-methyladenine (6 mA) (Wang et al., 2020). Methylation of cytidine-guanosine (CpG) dinucleotides of DNA mainly occurs at position 5 of cytosine to yield 5-methylcytosine (5mC), which is responsible for one-third of spontaneous mutations and ultimately the cause for cancer (Jin et al., 2013; Plongthongkum et al., 2014). DNA methylation depends on many factors, for example, the environment (as chemical agents), age, gender, the presence of other diseases, different cells, and the phase of the cell life (Wang et al., 2020).
DNA methylation has been linked to many human genetic diseases not only restricted to cancer but also autoimmune diseases, diabetes, neurodegenerative diseases, including Alzheimer’s and Parkinson’s diseases (Ehrlich, 2019; Kader et al., 2018) besides ageing and metabolic disorders (Wang et al., 2020). The presence of 5mC can cause gene silencing and consequently change binding protein transcription. Additionally, 5mC adds hydrophobicity to the DNA main groove, which in turn affects the functions of DNA-binding proteins (Carvalho et al., 2014).
This epigenetic mark is heritable and conserved during cell division (Kader et al., 2018). DNA methylation adds genetic information but does not change the base sequence of DNA. Since 5mC is also a complementary base for guanine, 5mC and cytosine are indistinguishable by sequencing (Jin et al., 2013). DNA methylation has a significant role in many biological processes, including embryonic development, regulating gene expression, inactivation of X-chromosome and gene silencing (Aranda et al., 2016; Bestor et al., 2015; Cotton et al., 2015; Fu and He, 2012; Jjingo et al., 2012; Kader et al., 2018; Li et al., 1992). Moreover, methylation reaction can lead to a pre-mutagenic site due to the formation of the thymine (T) upon the deamination of the 5mC, generating an error in base pairing (Bestor and Coxon, 1993; Bestor and Verdine, 1994). This is different from the deamination of Cyt that results in Uracil, as the latter can be repaired by Uracil-DNA glycosylases (UDGs) enzyme (Robertson and Wolffe, 2000). Deamination reactions of nucleic acid bases have been widely explored, and the findings are extremely useful in suggesting the processes to be addressed in this paper (Almatarneh et al., 2020, 2017a, 2017b, 2008, 2006; Alrawashdeh et al., 2013; Halim et al., 2014; Uddin et al., 2011).
According to the results of molecular dynamics (MD) simulations, the methyl group adds extra steric hindrance to the helix and reduces its flexibility. Hence, it affects the ability of the proteins to be wrapped around the helix (Severin et al., 2011).
The family of de novo DNA methyltransferases (DNMT) contains two important active enzymes, DNMT3A and DNMT3B, which are able to catalyze the methylation at the C5-position of cytosine with S-adenosyl-L-methionine (AdoMet) as a methyl donor (Aranda et al., 2010; Chua et al., 2019, p.). Recently, it has been proved experimentally that the DNMT enzyme has an off-target activity toward N3 Yeilding 3-methylcytosine (3mC) (Dukatz et al., 2019; Rošić et al., 2018). Most recently, we have investigated the reaction mechanisms of 3mC and performed DFT calculations to calculate the activation energy of this off-target activity (Almatarneh et al., 2022).The methylated one is preserved by DNMT1 through cell division and replication (Carvalho et al., 2014). The family of DNMTs comprises two main parts in their structures, known as the N-terminal and the C-terminal. The two parts are structurally similar between eukaryotes and prokaryotes. The C-terminal embraces the active site of ten amino acid motifs and a core structure named “AdoMet-dependent MTase fold” that facilitates the transfer of methyl group to cytosine in DNA. DNMTs follow the general mechanism of enzymes by flipping the target nucleic base out of the DNA helix into the active site pocket (Jurkowska et al., 2011; Klimasauskas et al., 1994).
The mechanism for the methylation of cytosine at the C5 position is outlined in Scheme 1. At first, the cytosine ring is subjected to a nucleophilic attack at C6 by a thiolate ion (RS¯) provided by the cysteine residue of an enzyme. The nucleophilic attack leads to the formation of a DNA-enzyme adduct, in which the C5 position is activated as a nucleophile to subsequently attack an electrophilic methyl donor (AdoMet), resulting in the methylation of the C5 position. Finally, the methylated adduct undergoes a base-promoted elimination step to give 5mC as the final product.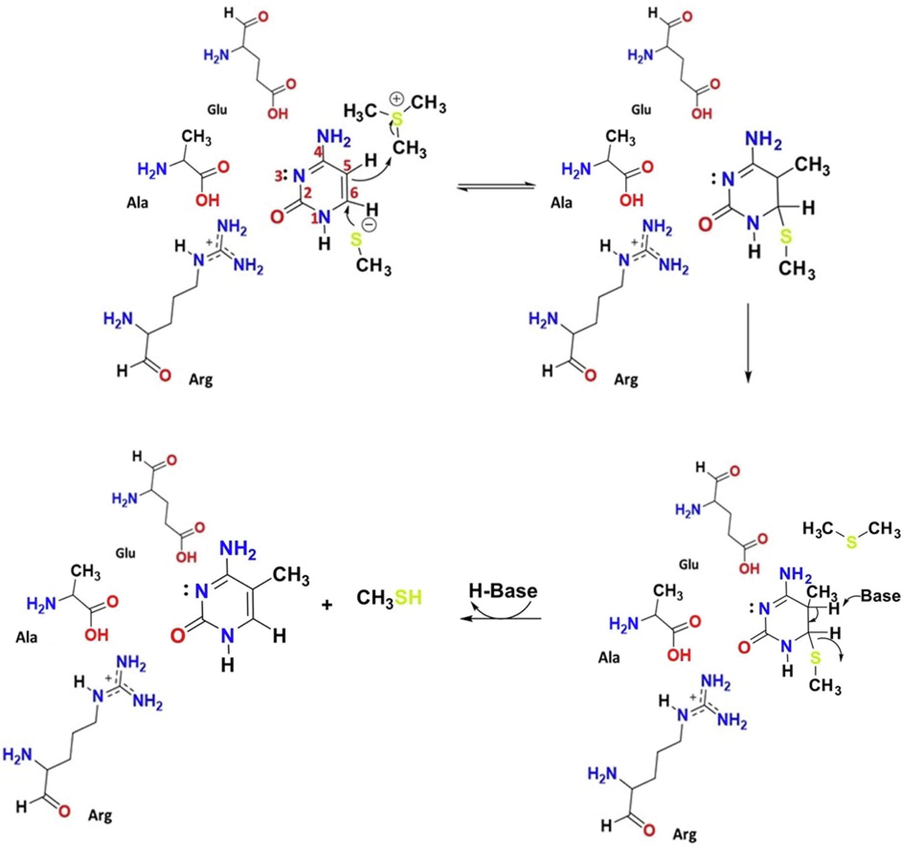
Mechanism of cytosine methylation by DNMT.
This methylation mechanism has already been investigated by various groups through theoretical and experimental approaches. Peräkylä carried out ab initio (up to the MP2/6–31 + G(d)//HF/6–31 + G(d)) levels of theory) and density functional theory (DFT) calculations (B3LYP/6–31 + G(d) level of theory) to investigate a non-concerted methylation mechanism in both aqueous and gaseous phases. The roles of protonation and deprotonation of N3 in catalyzing the reaction were examined through calculations of the pKa values of N3 of the ring. Their calculations showed that methylation of the activated C5 occurs fast and is exothermic. The enzyme DCMtase accelerates the methylation process by placing the reactive group in a way that involves a proton transfer from the Glu119 to N3 of cytosine. For the step of product formation, the methyl thiolate residue of Cys81-S¯ (Cysteine) was proposed to undergo a syn elimination by changing the ring conformation of C6-S and C5-H5 bonds into a syn-periplanar conformation to facilitate concerted H5 and Cys81-S¯ elimination (Peräkylä, 1998).
Through a QM/MM approach, Zhang et al. (Zhang and Bruice, 2006) illustrated the concerted uncatalyzed reaction of the nucleophilic attack of Cys-81-S¯ to C6 and AdoMet to C5 in an aqueous medium. The calculated activation energy of this reaction was found to be 34.7 kJ mol−1. The roles of Glu119, Arg163, and Arg165 were studied, and they concluded that Glu119 lacks catalytic activity, while Arg163 and Arg165 give a minor electrostatic stabilization. They also examined hydroxide ion (OH–) as the base to abstract the H5 proton of the cytosine-enzyme adduct. The activation energy of this step was around 50.2 kJ mol−1 at pH 7.
Another independent study by Yang et al. (Yang et al., 2013) analyzed the methylation reaction by QM/MM-MD calculations, where the role of Glu 119 was studied. They concluded that the nucleophilic attack of Cys81-S¯ at C6 and methyl addition at C5 occur concertedly with the aid of Glu119 residue, which protonates cytosine N3 position. The calculated free energy of activation of this step was 66.1 kJ mol−1. Elimination of the C5 proton was the rate-determining step. OH– base is present in the active site through a proton wire from the bulk water, and the barrier for this step was 86.6 kJ mol−1 (Yang et al., 2013).
Aranda and co-workers utilized QM/MM-MD to study the methylation reaction; the concerted first step was found to overcome a barrier of 79.9 kJ mol−1. They concluded that protonation by Glu119 at N3 is not required at this stage, but is required in the next step to faciliate the proton abstraction by H2O molecule with an activation energy of 78.2 kJ mol−1 (Aranda et al., 2016).
Most recently, Jerbi and co-workers used a combined method of MD and DFT, similar to that by Aranda (Aranda et al., 2016), to assess the methylation reactions in DNA. They investigated the non-concerted nucleophilic activation by cysteine residue, followed by the methyl addition with methyl donors, including AdoMet and other similar methyl donors deposited in the PubChem database. The free energy of activation, when AdoMet was used as the methyl donor, was calculated as 56.7 kJ mol−1 (Jerbi et al., 2017).
Despite the existing of theoretical studies, some mechanistic details are still controversial and need further clarification concerning the roles of the glutamic acid residue, the water molecules around the active site, and the base that abstracts the proton during the β-elimination step.
In this study, we performed a computational study based on the most plausible reaction mechanism shown in Scheme 1. Aiming at determining the role of water molecules present in the active site, as it is well known that water has a crucial impact on the biological systems, we utilized water as a bridge to transfer the H5 proton. We also investigated the debatable role of glutamic acid (Glu) residue by mutating it into alanine (Ala) amino acid in the presence of other residues, such as arginine (Arg) and cysteine (Cys). Based on the results of our DFT calculations, mechanistic details concerning the reactivity of DNMTs towards C5 of cytosine in the gaseous and aqueous phases have been established.
2 Computational method
Geometry optimizations were conducted at the B3LYP/6-31G(d), B3LYP/6–31 + G(d), B3LYP/6-31G(2df,2p) levels of theory using the Gaussian 16 (G16) quantum chemistry package. The optimized geometries were confirmed to be either energy minima or transition states (TSs) on the potential energy surface by calculating their vibrational frequencies at the same levels of theory used for optimization. An energy minima geometry does not show any imaginary frequencies, while a TS shows only one imaginary frequency. Furthermore, TSs were subjected to intrinsic reaction coordinate (IRC) calculations at the B3LYP/6-31G(d) level of theory in order to connect the TSs with the local minima of the reactants and products on the potential energy surface. The proposed reaction pathways were studied in the aqueous medium using the solvation model based on density (SMD) at the B3LYP/6–31 + G(d) level of theory. Thermodynamic parameters: ΔS, ΔH, and ΔG, as well as the kinetic parameters, including activation energy (Ea), enthalpy of activation (ΔH‡), and the Gibbs energies of activation (ΔG‡) were calculated for each proposed reaction pathway.
3 Results and discussion
In this study, comprehensive quantum chemistry calculations were conducted to investigate the mechanisms for the two main reaction pathways proposed for the methylation reaction of cytosine at position 5.
The role of water molecules present in the active site during the methylation reaction were examined. According to energy minimization with the QM/MM Hamiltonian obtained for the ground state active site of M. HaI (bacterial M. HaI DNA methyltransferase), which shares the same methylation mechanism with mammalian enzyme) (Yang et al., 2013), there are four water molecules surrounding the flipped cytosine ring. The N3 of cytosine is H-bonded to Wat-330 at a distance of 2.89 Å. Wat-353 is H-bonded to H5 at 2.45 Å, and Wat-340 is H-bonded to O2 of cytosine at 1.64 Å (see Fig. 1 for details). A number of reaction schemes that take one, two and three water molecules into account, were investigated in our studies. A maximum of two water molecules were considered to take part in H5 bridging. Herein, we report the best results that involve one water molecule only.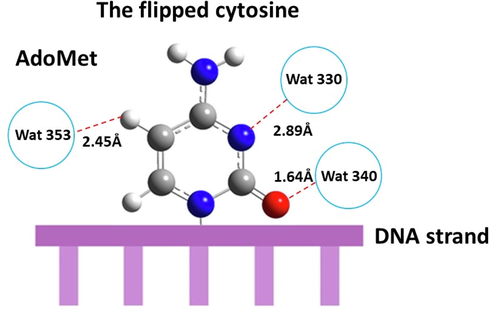
The target cytosine ring and the surrounding water molecules, Wat-353, Wat-330 and Wat-340. Their corresponding H-bonds to the ring are indicated.
Our pathways were designated to also include the other active site residues. Pathway A contains Glu amino acid, Arg, and Cys as shown in Scheme 2A, while in pathway B Glu is mutated into Ala, as depicted in Scheme 2B below.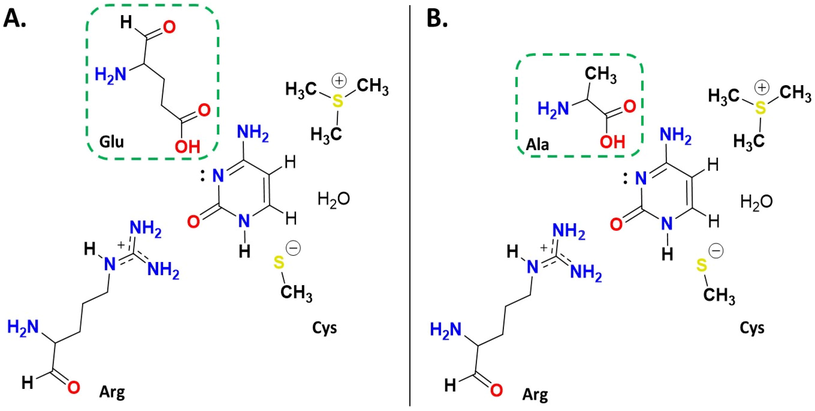
Schematic diagram illustrating our proposed pathways: A. pathway A and B. pathway B. Glu is mutated in pathway B to Ala.
The activation energies (Ea), enthalpies of activation (ΔH‡), and Gibbs energies of activation (ΔG‡) were calculated at different basis sets for our proposed pathways and provided in Tables 1 and 2. Moreover, the potential energy diagrams (PEDs) were constructed and displayed in Fig. 4 and Fig. 7 at different basis sets in aqueous and gaseous phases (in kJ mol−1 at 298.15 K). The thermodynamic parameters for the proposed pathways are presented in Table 3.
Basis set
TS1A
TS2A
Ea
ΔG‡
Ea
ΔG‡
B3LYP/6-31G(d)
54
55
157
168
B3LYP/6–31 + G(d)
47
48
163
179
B3LYP/6-31G(2df,2p)
51
33
154
164
SMD*
49
45
142
151
Basis set
TS1B
TS2B
Ea
ΔG‡
Ea
ΔG‡
B3LYP/6-31G(d)
31
37
161
171
B3LYP/6–31 + G(d)
44
44
166
178
B3LYP/6-31G(2df,2p)
37
44
173
176
SMD*
46
46
161
160
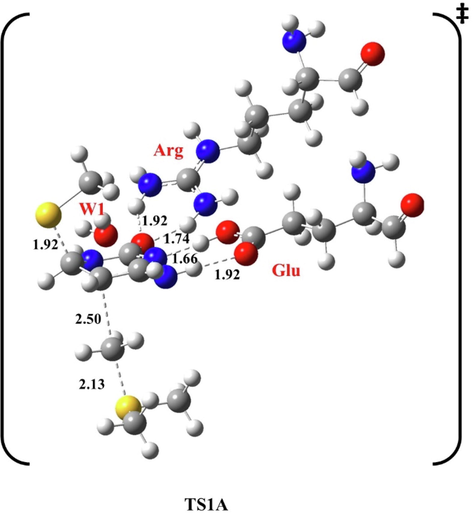
Optimized structure of the first transition state (TS1A) of methylation reaction, at B3LYP/6-31G(d), pathway A. Bond lengths are in Å.
Pathway
B3LYP/6-31G(d)
B3LYP/6–31 + G(d)
B3LYP/6-31G(2df,2p)
SMD*
ΔH
ΔG
ΔH
ΔG
ΔH
ΔG
ΔH
ΔG
A
−230
−256
−221
−243
−229
−267
−173
−196
B
−258
−282
–233
−256
−248
−274
−163
−200
3.1 Methylation reaction in the presence of Glu: Pathway a
In our study, the concerted mechanism of the methylation reaction was analyzed. In the first step, methyl thiolate initiates a nucleophilic activation at C6, and CH3 is transferred to C5. The second step involves the β-elimination of H5 by an H2O bridge molecule, (see Scheme S1 in Supplementary Material). In the first transition state (TS1A), the flipped cytosine was stabilized by H-bonding between glutamic acid (Glu) and H4 at 1.92 Å and 1.66 Å from N3, while Arg forms two H-bonds with O2 at 1.74 Å and 1.92 Å, respectively.
W1 was on the other side of the target ring at 2.30 Å from H4 and 2.46 Å from H5, as shown in the optimized geometries of the first transition state at B3LYP/6-31G(d) level of theory (Fig. 2). Methyl thiolate attacks C6 at 1.92 Å, and CH3 approaches C5 at 2.50 Å.
In TS2A, Glu forms H-bonds with N3 at 1.78 and 1.85 Å with H4. H5 moves away from C5 at 1.46 Å to W1 at 1.18 Å, then from W1 at 1.11 Å to the cysteine residue at 1.80 Å. However, Arg is at 1.77 Å and 2.05 Å from O2.
The resulting methyl thiol departs at 2.52 Å from C6, and the double bond between C5 and C6 is formed at 1.45 Å, as indicated in Fig. 3.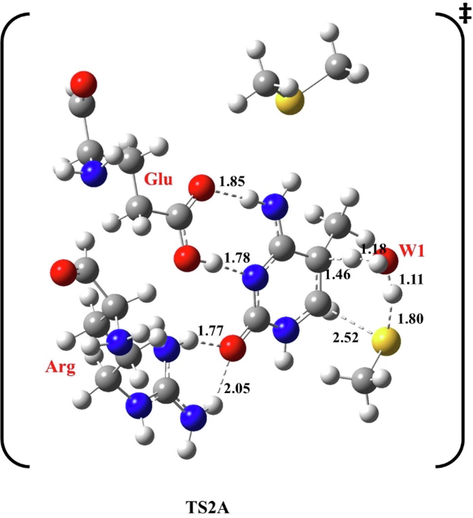
Optimized structure of the first transition state (TS2A) of methylation reaction, at B3LYP/6-31G(d), pathway A. Bond lengths are in Å.
The activation energy of the first step of pathway A was calculated at different basis sets, as shown in Table 1 and Fig. 4.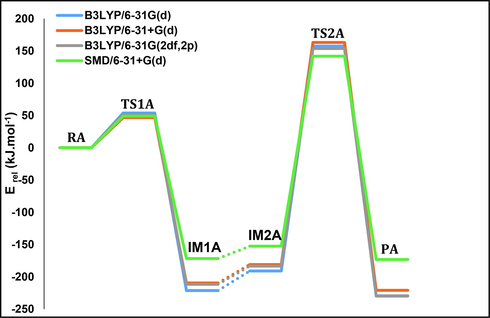
The potential energy diagram of pathway A calculated at different basis sets in aqueous and gaseous phases, in kJ mol−1 at 298.15 K.
For TS1A, the activation energy calculated at the B3LYP/6-31G(d) level is 54 kJ mol−1. In an aqueous environment it is 49 kJ mol−1 based on calculations done with SMD/6–31 + G(d). The results obtained from other basis sets are highly correlated and consistent (see Table 1). Our calculated activation energy is in good agreement with previously reported values and close to the experimental value, 43.1 kJ mol−1 (10.3 Kcal mol−1) (Aranda et al., 2016; Jerbi et al., 2017; Sharma et al., 2005; Yang et al., 2013).
For the second step, the activation energy for TS2A is 157 kJ mol−1 at the B3LYP/6-31G(d) level of theory, and in an aqueous environment it is 142 kJ mol−1 using SMD/6–31 + G(d). The final methylation product has an energy of −230 kJ mol−1, which is the most stabilized among other studied pathways.
3.2 Methylation reaction in the presence of Ala: Pathway B
In this pathway, we carried a methylation reaction assuming the existence of Ala, Arg, and one water molecule while mutating Glu. The motivation behind this designation was to detect if the Glu has catalytic activity, besides investigating the catalytic role of Ala.
In this pathway, the same general mechanism described in the previous sections is followed. In TS1B, Ala forms a H-bond with N3 at 1.65 Å, while Arg forms two H-bonds with O2 at 1.87 Å and 2.00 Å, respectively, as shown in Fig. 5.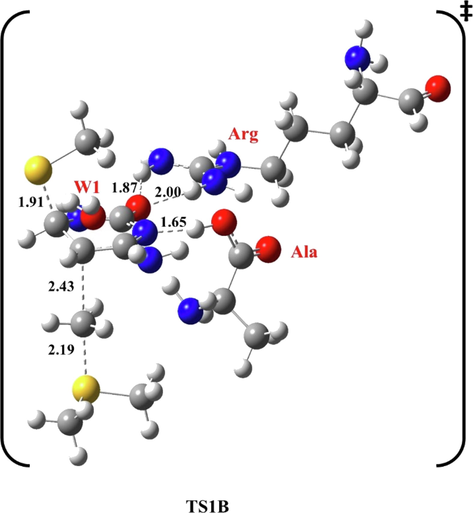
Optimized structure of the first transition state (TS1B) of methylation reaction, at B3LYP/6-31G(d), pathway B. Bond lengths are in Å.
W1 is on the other side of the target ring at 2.14 Å from H4 and 2.50 Å from H5, of the first transition state at B3LYP/6-31G(d) level of theory. Methyl thiolate attacks C6 at 1.91 Å, and CH3 approaches C5 at 2.43 Å.
In TS2B, Ala becomes in the ring plane, forming H-bonds with N3 at 1.83 and 1.99 Å with H4 and 2.07 Å from Arg. However, Arg is at 1.90 Å and 2.08 Å from O2. The H5 proton left C5 at 1.46 Å to W1 at 1.19 Å, then from W1 at 1.12 Å to the cysteine residue at 1.78 Å. The resulting methyl thiol departed at 2.53 Å from C6, and the double bond between C5 and C6 is rebuilt at 1.45 Å, see Fig. 6.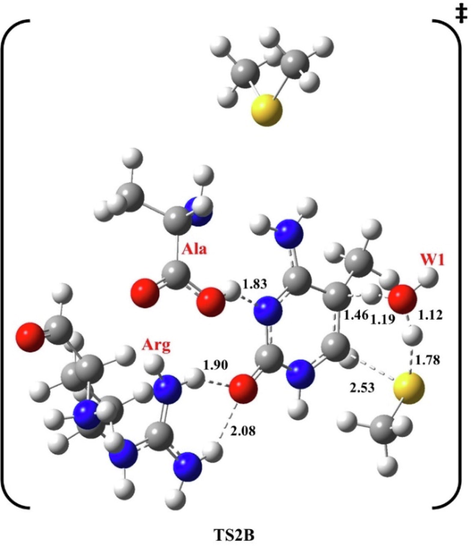
Optimized structure of the second transition state (TS2B) of methylation reaction, at B3LYP/6-31G(d), pathway B. Bond lengths are in Å.
The activation energy of the first step of pathway B was calculated at B3LYP/6-31G(d) as shown in Table 2 and Fig. 7.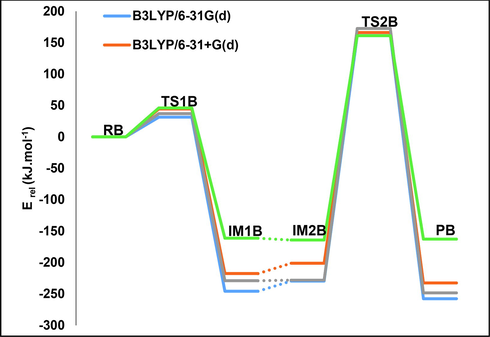
The potential energy diagram of pathway B calculated at different basis sets in aqueous and gaseous phases, in kJ.mol−1 at 298.15 K.
Our results obtained by other basis sets are consistent with each other. For TS1B, the activation energy is 31 kJ mol−1 calculated at the B3LYP/6-31G(d) level of theory, and in the aqueous environment utilizing SMD/6–31 + G(d) it is 46 kJ mol−1. Our calculated activation energy is close to the experimental value mentioned previously, 43.1 kJ mol−1 (10.3 Kcal.mol−1) (Sharma et al., 2005).
For the second step, the activation energy for TS2B is 161 kJ mol−1 at the B3LYP/6-31G(d) level of theory and in the aqueous environment applying SMD/6–31 + G(d) it is 46 kJ mol−1. The final methylation product has a relative energy of −258 kJ mol−1, which is the most stabilized among other pathways studied.
Comparing our obtained results of pathway B, we conclude that the activation energy of pathway B is the nearest to the experimental value meant for the first step, in addition to merging the role of the vital residues of the active site, such as Ala. This is perfectly consistent with the experimental observation that concluded the impact and importance of Ala residue in this reaction by replacing residues of the active site with Ala in particular (Sharma et al., 2005). On another sight, Ala has fortunately proved its catalytic activity toward the formation of 3mC as explored in our previous work (Almatarneh et al., 2022). Ala has been reported to catalyze the methylation at N3 and lower the activation energy of this reaction. We believe that our results are coherent and emphasize the significant impact of both methylation patterns (3mC and 5mC).
3.3 Thermodynamic parameters for cytosine methylation at C5.
The thermodynamic parameters (ΔH and ΔG) for the proposed pathways of methylation reaction of cytosine at position 5 are given in Table 3 at all studied basis sets. The methylation reaction of cytosine at position 5 in pathway A and in the presence of amino acids Glu and in the presence of Ala in pathway B are found to be exothermic and exergonic in aqueous and gaseous phases at all basis sets. According to the results obtained in Table 3, pathway B has the lowest thermodynamic parameter values. Therefore, it is more spontaneous and plausible reactions to occur.
4 Conclusion
In this work, we have conducted a mechanistic exploration of the methylation at the C5 position of the cytosine ring utilizing quantum chemical DFT calculations. The role of conserved residues, for instance, alanine (Ala), glutamic acid (Glu) and arginine (Arg) and abundant water molecules were considered in two pathways A and B for this reaction, where Glu is mutated to Ala in pathway B in order to determine the catalytic role of both residues on the reaction.
Herein the reaction occurs via two main steps; the first involves a concerted nucleophilic addition of methyl thiolate at C6 and methyl transfer at C5. Followed by the second step of H5 elimination by H2O molecule through bridging the H5 to the departed methyl thiolate.
Our results address that alanine and arginine can benefit C5 methylation reaction by reducing the activation energy, while Glu shows an insignificant catalytic activity. The most feasible pathway for 5mC would be the one containing Ala, Arg, and one water molecule with an activation energy of 31 kJ mol−1. We believe that these results can contribute to the understanding of the role of active site residues.
Acknowledgements
I have acknowledged the fund that was provided from the Deanship of Academic Research at the University of Jordan and there is no grant ID that can be used.
References
- Mechanistic and spectral investigation on the deamination of ammeline and ammelide. Comput. Theor. Chem.. 2017;1117:92-99.
- [CrossRef] [Google Scholar]
- A computational mechanistic study of the deamination reaction of melamine. Int. J. Quantum Chem.. 2017;117:180-189.
- [CrossRef] [Google Scholar]
- Mechanisms for the Deamination Reaction of Cytosine with H2O/OH− and 2H2O/OH−: A Computational Study. J. Chem. Inf. Model.. 2008;48:831-843.
- [CrossRef] [Google Scholar]
- Computational Study of the Deamination Reaction of Cytosine with H2O and OH-. J. Phys. Chem. A. 2006;110:8227-8234.
- [CrossRef] [Google Scholar]
- Computational Insights in DNA Methylation: Catalytic and Mechanistic Elucidations for Forming 3-Methyl Cytosine. J. Chem.. 2022;2022:2673396.
- [CrossRef] [Google Scholar]
- Almatarneh, M.H., Omeir, R.A., AL Demour, S., Elayan, I.A., Islam, S., Poirier, R.A., 2020. Hydrolytic deamination mechanisms of guanosine monophosphate: A computational study. Computational and Theoretical Chemistry 1175, 112732. https://doi.org/10.1016/j.comptc.2020.112732.
- Computational study on the deamination reaction of adenine with OH−/nH2O (n = 0, 1, 2, 3) and 3H2O. Can. J. Chem.. 2013;91:518-526.
- [CrossRef] [Google Scholar]
- Theoretical Study of the Catalytic Mechanism of DNA-(N4-Cytosine)-Methyltransferase from the Bacterium Proteus vulgaris. J. Phys. Chem. B. 2010;114:8467-8473.
- [CrossRef] [Google Scholar]
- Unraveling the Reaction Mechanism of Enzymatic C5-Cytosine Methylation of DNA. A Combined Molecular Dynamics and QM/MM Study of Wild Type and Gln119 Variant. ACS Catal.. 2016;6:3262-3276.
- [CrossRef] [Google Scholar]
- Cytosine methylation The pros and cons of DNA methylation. Curr. Biol.. 1993;3:384-386.
- [CrossRef] [Google Scholar]
- Notes on the role of dynamic DNA methylation in mammalian development. Proc Natl Acad Sci USA. 2015;112:6796.
- [CrossRef] [Google Scholar]
- Understanding the structural and dynamic consequences of DNA epigenetic modifications: Computational insights into cytosine methylation and hydroxymethylation. Epigenetics. 2014;9:1604-1612.
- [CrossRef] [Google Scholar]
- Cytosine-Based TET Enzyme Inhibitors. ACS Med. Chem. Lett.. 2019;10:180-185.
- [CrossRef] [Google Scholar]
- Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet.. 2015;24:1528-1539.
- [CrossRef] [Google Scholar]
- Mechanistic Insights into Cytosine-N3 Methylation by DNA Methyltransferase DNMT3A. J. Mol. Biol.. 2019;431:3139-3145.
- [CrossRef] [Google Scholar]
- DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics. 2019;14:1141-1163.
- [CrossRef] [Google Scholar]
- Nucleic acid modifications with epigenetic significance. Curr. Opin. Chem. Biol.. 2012;16:516-524.
- [CrossRef] [Google Scholar]
- Mechanistic Study of the Deamidation Reaction of Glutamine: A Computational Approach. J. Phys. Chem. B. 2014;118:2316-2330.
- [CrossRef] [Google Scholar]
- Jerbi, J., Springborg, M., den-Haan, H., Cerón-Carrasco, J.P., 2017. S-adenosyl-l-methionine analogs as enhanced methyl donors: Towards novel epigenetic regulators. Chemical Physics Letters 690, 74–81. https://doi.org/10.1016/j.cplett.2017.10.042.
- Effects of Protonation and C5 Methylation on the Electrophilic Addition Reaction of Cytosine: A Computational Study. J. Phys. Chem. B. 2013;117:3-12.
- [CrossRef] [Google Scholar]
- On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3:462-474.
- [Google Scholar]
- Structure and Function of Mammalian DNA Methyltransferases. ChemBioChem. 2011;12:206-222.
- [CrossRef] [Google Scholar]
- The effects of DNA methylation on human psychology. Behav. Brain Res.. 2018;346:47-65.
- [CrossRef] [Google Scholar]
- Hhal methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357-369.
- [CrossRef] [Google Scholar]
- Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915-926.
- [CrossRef] [Google Scholar]
- A Model Study of the Enzyme-Catalyzed Cytosine Methylation Using ab Initio Quantum Mechanical and Density Functional Theory Calculations: pKa of the Cytosine N3 in the Intermediates and Transition States of the Reaction. J. Am. Chem. Soc.. 1998;120:12895-12902.
- [CrossRef] [Google Scholar]
- Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat. Rev. Genet.. 2014;15:647-661.
- [CrossRef] [Google Scholar]
- Evolutionary analysis indicates that DNA alkylation damage is a byproduct of cytosine DNA methyltransferase activity. Nat. Genet.. 2018;50:452-459.
- [CrossRef] [Google Scholar]
- Cytosine methylation alters DNA mechanical properties. Nucleic Acids Res.. 2011;39:8740-8751.
- [CrossRef] [Google Scholar]
- Residues Distal from the Active Site that Alter Enzyme Function in M.HhaI DNA Cytosine Methyltransferase.. 2005;null 22:533-543.
- [CrossRef]
- Mechanistic Study of the Deamination Reaction of Guanine: A Computational Study. J. Phys. Chem. A. 2011;115:2065-2076.
- [CrossRef] [Google Scholar]
- Wang, L., Li, X., Tang, D., Wang, W., 2020. Chapter 7 - DNA methylation, in: Forero, D.A., Patrinos, G.P. (Eds.), Genome Plasticity in Health and Disease. Academic Press, pp. 93–108. https://doi.org/10.1016/B978-0-12-817819-5.00007-3.
- DNA Cytosine Methylation: Structural and Thermodynamic Characterization of the Epigenetic Marking Mechanism. Biochemistry. 2013;52:2828-2838.
- [CrossRef] [Google Scholar]
- Zhang, X., Bruice, T.C., 2006. The mechanism of M.HhaI DNA C5 cytosine methyltransferase enzyme: A quantum mechanics/molecular mechanics approach. Proceedings of national academy of sciences 103(16), 6148–6153. https://doi.org/10.1073/pnas.0601587103.
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.103956.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary Data 1
Supplementary Data 1







