Translate this page into:
Molecular analysis of recombinant collagenase from Bacillus siamensis strain Z1: Gene Cloning, expression and in-silico characterization
⁎Corresponding author at: Dr.Zabin K. Bagewadi, Associate Professor, Department of Biotechnology, KLE Technological University, Vidyanagar, Hubballi 580031, India. zabin@kletech.ac.in (Zabin K. Bagewadi),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
Cloning and expression of collagenase gene from Bacillus siamensis strain Z1. Biochemical characterization for enzyme profiling. Analysis included MALDI-TOF-MS, N-terminal sequencing, CD, 1H NMR and TGA. In-silico studies included characterization, Molecular docking, simulations and MMPBSA.
Abstract
This study focuses on gene cloning, expression, biochemical and analytical characterization along with structural and functional characterization of collagenase followed with molecular docking, dynamics study and Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA). The collagenase gene identified from the genome of the novel collagenase Bacillus siamensis strain Z1 is introduced into E.coli DH5α, subsequently expressed in E.coli BL21 (DE3) withisopropyl β −d – 1 − thiogalactopyranoside(IPTG) induction and further affinity purified yielding in ∼ 89.4 kDa recombinant collagenase which demonstrated alkali characteristics and thermostability determined by thermodynamic parameters. Recombinant collagenase revealed good stability when exposed to diverse biochemical components. The recombinant collagenase identity was confirmed through matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) showing specific mass peaks and via N-terminal sequence analysis as MTAVNQTISK. Moreover, the concluded N-terminal amino acid sequence from Edman degradation displayed significant resemblance. The structural and functional analysisof recombinant collagenase was analysed by Circular dichroism (CD), Proton nuclear magnetic resonance (1H NMR) spectrometry and Thermogravimetric analysis (TGA). The recombinant collagenase also showed gelatin liquefaction ability. The collagenase gene sequence is also assessed for structural and functional characterizations by using various computational tools and revealed its classification in U32 family peptidase. A Grand average of hydropathicity index (GRAVY) score of −0.295 and instability index of 37.22 was obtained. Homology model of collagenase gene was generated by employing SWISS-MODEL and structure analysis by Ramachandran plot. Molecular docking of modelled collagenase with four different substrates was carried out by PyRx and Autodocking. Highest docking score of −12.7 kcal/mol was obtained for Alaska pollock hydroxyproline containing marine collagen peptide (APHCP). Subsequently, Molecular dynamics and simulations for highest score docked complex was assessed using GROningenMAchine for Chemical Simulations (GROMACS).
Keywords
U32 peptidase
Molecular cloning
Purification
Biochemical and structural evaluation
Computational analysis
1 Introduction
The expanding fishery industry generates substantial collagen-rich waste yet inefficient management leads to underutilization (Bhagwat and Dandge, 2018). Collagen fibrous structure resists degradation. The trimeric helix of collagen consists of similar or different polypeptide chains supercoiled counterclockwise around an axis, forming a single triple helix with glycine atoms. Collagenases are a class of hydrolytic proteases that can break down different kinds of collagen. The primary sources of collagenases include microorganisms, plants and animals with varying substrate specificities. Microbial collagenases (EC 3.4.24.3) possess extensive substrate specificity that aids in breaking down both water-insoluble and water-soluble forms of collagens within the triple helical regions at X-Gly bonds (Pal and Pv, S, 2016). Marine collagenases excel in catalyzing marine collagen, especially from fish, attracting interest due to their exceptional qualities (Yang et al., 2017). Microbial collagenases consist of matrix metalloproteases (MMP)that comprise of collagenases (MMP-1, MMP-8, MMP-13 and MMP-18) that can break down four types of collagens (I, II, III, IV) (Alipour et al., 2016). Serine proteases from families S8, S1, and S53 might also fall into collagenases. Keratinases, a new class of proteases, hydrolyze tough keratins and are classified within the serine and metalloprotease families, as documented in the MEROPS database (Rawlings et al., 2018). They primarily fall under family S8 of clan SB, which encompasses enzymes like trypsin and subtilisin. Furthermore, certain U32 family members are also recognized as collagenases (Bhagwat and Dandge, 2018). Not many reports of collagenase mechanism of Bacillus sp. are reported, Vibrio collagenase hydrolyzes collagen using peptidase M9N and pre-peptidase C-terminal domains, breaking down the C – telopeptide region and fragmenting tropocollagen fragments into peptides and amino acids, primarily at Y-Gly bonds(Wang et al., 2022).
Microbial collagenases have numerous industrial applications, including wound healing, placenta treatment, and therapeutic applications like Dupuytren’s disease, Peyronie’s disease and Glaucoma(Alipour et al., 2019). They also produce bioactive collagen peptides with antioxidant, antimicrobial, anticancer, antidiabetic, and anti-inflammatory properties(OLIVEIRA et al., 2017; Yang et al., 2017;Nurilmala et al., 2020; Lima et al., 2015; Kumar et al., 2019). They are crucial in scientific research and food industry(Bhagwat and Dandge, 2018).
Researchers have modified collagenase through genetic engineering to improve its expression, purifying microbial collagenases from selected bacterial species and sequencing associated genes. (Pal and Pv, S, 2016). Molecular cloning with suitable vectors is a suitable method for obtaining collagenase, as native collagenases can negatively impact biosynthesis due to lack of firmness and different iso-enzymatic configurations. (Song et al., 2021). Recombinant collagenase is constructed using tags like, His tag and MBP tag. MBP tag improves the solubility of the target protein by aiding in correct folding and leads to subsequent increase in yield (Dutta and Bose, 2022). Expression systems like E.coli and Brevibacillus are used for successful collagenase gene expression(Zhu et al., 2022; Teramura et al., 2011). Using amylose affinity chromatography recombinant proteins have been purified. Through biochemical characterization of recombinant collagenase it is important in evaluating their suitability across various fields where they are intended for practical utilization(Bhagwat and Dandge, 2018).
In today’s scientific research, various in-silico tools are utilized to analyze protein structures, exploration of folding patterns, identify structural elements and family domains. 3D structures are predicted and elucidate functional features (Rani and Pooja, 2018). In-silico studies enable protein engineering for various biotechnological applications. Molecular docking predicts interactions between proteins and ligands, aiding in drug development and enzyme substrate binding. Advancements in in-silico techniques involve molecular dynamics and simulations, providing insights into dynamic behavior, interactions, and structural changes. This computational approach helps to understand biological processes and explore molecular mechanisms(Revankar et al., 2023; Mechri et al., 2022; Bhatt et al., 2021).
The cloning and expression of the collagenase gene from Bacillus siamensis strain have not been previously reported, and there is limited analytical characterizations available and very rare reports on structural, functional characterization of collagenases from Bacillus sps., and their molecular docking and dynamics studies with substrates indicates limited prior research and novelty in this area. The current research identified collagenase gene, cloned it using a pET28 vector, and expressed it in E. coli. subsequently, purification and biochemical characterization of recombinant collagenase gene yielded a molecular weight of ∼ 89.4 kDa. Various analytical methods were used, including matrix assisted laser desorption ionization-time of flight mass spectrometry(MALDI-TOF-MS), Circular Dichroism (CD),Nuclear Magnetic Resonance spectroscopy (NMR), and Thermogravimetric Analysis(TGA), while computational tools were employed to study its structural and functional characteristics, molecular docking and stability with Alaska pollock hydroxyproline containing marine collagen peptide(APHCP) substrate.
2 Materials and methods
2.1 Chemicals and vectors
The chemicals and resins utilized in the current study was sourced through reputable suppliers, including Sigma-Aldrich Co. and Merck and Co., Inc. (USA). The collagenase gene was identified from Bacillussp. strain Z1 and cloning, expression and sequencing method was conducted. Molecular cloning and expression procedures involved the utilization of E. coli strains DH5α and E. coli BL21 (DE3), along with the pET28b (His Tagged) vector and pET28(Maltose Binding Protein (MBP) Tagged) vector. All reagents employed in this research were of research grade and readily accessible from marketable sources.
2.2 Identification of gene of interest
The quest to identify the gene of interest initiated with a nucleotide Basic Local Alignment Search Tool (BLAST) analysis of the 16S rRNA sequence, in tandem with the submission of the bacterial strain’s nomenclature. This analysis was conducted to uncover closely related sequences within BLAST search results. Subsequently, the BLAST outcomes were reviewed and selecting the accession number displaying the highest degree of sequence identity (Clough and Barrett, 2016). Sourced with this information, it was further accessed the respective National Center for Biotechnology Information (NCBI) whole genome sequences, which are customized to display all genetic features. Within this customized genomic context, the option to explore the entire genome sequence was selected that enabled to find and locate collagenase gene. This intricate process was iterated across multiple bacterial strains within the same species, ensuring a comprehensive survey. Furthermore, CLUSTAL Omega Multiple Sequence Alignment (MSA) Tool was employed to elucidate the degree of genetic correspondence between the collagenase gene sequences and that of strain Z1. This tool facilitated a systematic MSA of the collagenase gene sequences, thereby spotlighting collagenase genes that exhibited remarkable similarity to the one in strain Z1.
2.3 Primer designing, isolation of genomic DNA, amplification and cloning
Gene sequence primers are carefully designed with Snapgene Tool, optimizing the guanine-cytosine (GC) content and length. To ensure precise restriction digestion, BamHΙ and NotΙ restriction enzymes were utilized. The primers were strategically designed to include the respective restriction sites (BamHΙ – GGATCC; NotΙ – GCGGCCGC), positioned ahead of the primer sequences (Forward primer – 5′- AAAGGATCCATGACAGCCGTAAATCAAAC; Reverse Primer – 5′- AAAGCGGCCGCTTACTTCCCCTTTCTCATC). The genomic DNA (gDNA) was isolated from strain Z1 using Phenol Chloroform Extraction method. The collagenase gene was been amplified from genomic DNA utilizing specific designed primers mentioned above through Polymerase Chain Reaction (PCR). Subsequently, standard protocols were employed for conducting restriction digestion. The PCR procedure was performed utilizing a thermocycler and a Q5 High Fidelity DNA Polymerase with a reaction mixture volume of 50 µl. The protocol commenced with an initial denaturation step, which lasted for 2 min 30 sec at 98 °C. This was followed by denaturation at 98 °C for 20 sec by 35 cycles. Subsequently, primer annealing was subjected at hybridization temperature for a duration of 30 sec. the extension step followed, lasting 1 min 50 sec at 72 °C. The reaction was completed with a final extension step of 3 min at 72 °C. The resultant PCR products were then subjected to purification using purification kit(Bagewadi et al., 2016; Alei et al., 2023). The plasmid pET28 (MBP Tagged) was digested using restriction enzyme and then the plasmid and digested target gene were ligated and transformed into host E. coli DH5α competent cells by Heat shock method and further screened on Luria-Bertani (LB) agar plates supplemented with antibiotic Kanamycin. PCR analysis was performed on the recombinant plasmid employing primers specific to the plasmid in order to confirm the insertion and accurate alignment of target gene inside the vector and is confirmed by Gel Electrophoresis (Alei et al., 2023). To validate the accuracy of cloning, the isolated recombinant plasmid DNA obtained from the identified positive colonies was subjected to DNA sequencing. Further the plasmid was introduced into E. coli BL21 (DE3) cells using heat shock method, thereby facilitating the integration of plasmids within the cells. Colonies that showed successful transformation were then cultivated in LB media and collagenase protein expression was evaluated.
2.4 Collagenase expression by isopropyl β-D-1-thiogalactopyranoside (IPTG) induction
New colony of the transformed pET28 (MBP Tagged) vector was selected and cultivated on LB agar plate with E. coli BL21 competent cells. Initial culture was formed by introducing the culture into sterile LB medium (120 ml) supplemented with Kanamycin (50 µg/ml). The inoculated media was kept for overnight incubation at 37 °C under constant agitation at 180×g. The 120 ml starter culture was mixed with flask consisting 5 L of sterile LB medium contained Kanamycin and incubated at 37 °C under constant agitation (200 × g) for 4 h. Biomass concentration was checked to reach an optical density (OD) (600 nm) value of 0.55 – 0.7. Further 1 mM of IPTGwas been added to 4 h culture for inducing the protein expression and then the broth was incubated at 16 °C for 16 h under agitation condition (180 × g). Induced cell culture broth was centrifuged at 4500 × g, 4 °C for 45 min and the obtained cell pellets were preserved at −80 °C. Cell pellet was suspended again in chilled lysis buffer (0.5 mM NaCl, 10 mM Tris, 2.5 % v/v glycerol, and 0.2 % v/v Triton) and further to this 20 µL of benzamidine, 80 mg of lysozyme and 20 µL of phenylmethylsulfonyl fluoride (PMSF) is added and sonicated for 1 min at 60 amplitude for 6 cycles. The sonicated cells were centrifuged at 6000 × g at 4 °C for 45 min. Supernatant obtained was been recentrifuged at 13000 × g for 30 min at 4 °C to separate any debris and obtain a clean supernatant possessing genetically altered collagenase protein and is further assayed for collagenase activity as mentioned below. Purification by affinity chromatography was been carried using clear supernatant(Bagewadi et al., 2016; Alei et al., 2023).
2.5 Affinity purification, determination of molecular weight and enzymatic assay of genetically modified collagenase
Affinity chromatography was employed to purify recombinant collagenase using Amylose resin as solid matrix. The column loaded with resin was first washed with distilled water, then the column with amylose resin was pretreated with lysis buffer composing 0.5 mM NaCl, 10 mM Tris, 2.5 % v/v glycerol, and 0.2 % v/v Triton. Centrifugation was employed to obtain clear supernatant and it was mixed amylose resin in buffer followed by loading into column. Subsequently, the column underwent rinsing with wash buffer (50 mM Tris, 0.5 mM NaCl and 2.5 % Glycerol) to remove unbound proteins and contaminants effectively. The elution of the MBP-tagged protein was achieved by applying an elution buffer (50 mM Tris, 0.15 mM NaCl, 10 mM Maltose and 2.5 % Glycerol). Protein concentration at 280 nm was measured in order to record protein elution profile. After affinity chromatography, 12.5 % sodium dodecyl sulphate polyacrylamide gel electrophoresis was used to assess the purity of recombinant collagenase. Bradford assay with Bovine serum albumin (BSA) as standard was carried out to determine the recombinant collagenase concentration by measuring absorbance at 595 nm (Yaraguppi et al., 2022b). Collagenase activity was carried out according to Tran and Nagano (2002), with slight modifications. The reaction mixture was prepared by adding 1 ml of recombinant collagenase to 1 ml of collagen (marine fish collagen) (10 mg/ml) prepared in 50 mM Tris HCl buffer, supplemented with 4 mM CaCl2, pH 7.5. The reaction was performed for 30 min at 30 °C with agitation to suspend insoluble collagen and halted by adding 1 ml of 0.1 M acetic acid. Followed by centrifugation at 10,000 × gfor 15 min the quantity of liberated amino groups from the collagen was determined using Ninhydrin method. The absorbance was recorded at optical density 600 nm. The expression of collagenase activity is quantified as the µmol of leucine equivalent per min/ml of the culture (Tran and Nagano, 2002).
2.6 Biochemical characterization of purified recombinant collagenase
2.6.1 Determination of optimum pH and stability
The optimal pH of the recombinant collagenase has been ascertained by using different buffers at different pH levels previously outlined in the study conducted by Zhu et al. (2022), Alei et al., 2023. Using collagen as substrate, the effects of pH on collagenase activity was been examined in the pH range of 3––12 at 80 °C. The buffers included glycine – HCl (pH 3–4), sodium citrate buffer (pH 5–6), Tris – HCl (pH 7–9) and glycine NaOH (pH 9–12). To ascertain the pH stability, the enzyme was incubated for 20 h at 35 °C at pH 9 and pH 10. Samples were collected at fixed time intermittent and residual collagenase activity were measured, reported as relative collagenase activity.
2.6.1.1 Determination of optimal temperature and thermostability
The optimal temperature of recombinant collagenase was evaluated in selected temperature range from 35 °C to 80 °C at pH-9 (glycine – NaOH buffer) for 30 min using collagen as substrate. The control was collagenase activity without any additives. While for thermostability assessment of collagenase was done at regular intermittent of 2 h by incubating at temperature range of 50 – 60 °C for 20 h. Residual activity was measured at regular intervals and expressed as relative collagenase activity (Alei et al., 2023).
2.6.1.2 Effect of metal ions, chemicals, organic solvents and additives
The purified recombinant collagenase was used for its characterization. The study was assessed for effect of various metal ions (FeSO4, MgSO4, NaCl, KCl, CaCl2, ZnSO4, CuSO4 and MnSO4 at 0.5 M) and some additives (Urea, β- mercaptoethanol, dithiothreitol (LD-DTT) and ethylenediaminetetraacetic acid (EDTA) at 1 M). The evaluation involved the incubation of 100 µL collagenase with 200 µL of respective metal ion or additive for 4 h at 37 °C(Joshi and Satyanarayana, 2013). The surfactants effects were assessed by incubation of collagenase (100 µL) with 1 % of sodium dodecyl sulfate (SDS), Tween 80 and Triton X-100 respectively for 4 h at 37 °C (Wanderley et al., 2020). Collagenase stability with organic solvents (methanol, ethyl acetate, benzene, chloroform at 10 %) was been examined by incubating collagenase (100 µL) for 4 h at 37 °C with respective solvent (200 µL) (Joshi and Satyanarayana, 2013).
2.6.1.3 Thermodynamic analysis of collagenase
Thermodynamic assessment of the recombinant collagenase was conducted in accordance with the methodologies and formulas described by Abdella et al., 2023 and Mechri et al., 2022. The assessment of collagenase thermal deactivation was based on the specific equation:
Ln (Kd) and Ed (deactivation energy) were derived from the Arrhenius plot. The half-life (t1/2) of collagenase represents the time needed for a 50 % reduction in activity. The reduction time (D) indicates the time required for a 90 % reduction in activity at a given temperature range (70–90 °C) and can be calculated using the following formula:
The parameters associated with the thermal denaturation of collagenase, including thermodynamic denaturation (Kd), alterations in denaturation enthalpy (ΔHd), denaturation gibbs free energy (ΔGd) and denaturation entropy (ΔSd) were determined using the equations outlined in prior descriptions(Mechri et al., 2022).
2.6.1.4 Substrate specificity
The assessment of the substrate specificity of the recombinant collagenase involved testing its activity with a range of substrates. These substrates included collagen, gelatin, BSA, casein, azo-casein and hemoglobin. The collagenase reactions with each of these substrates followed standard assay procedures previously outlined (Tran and Nagano, 2002). In case of collagen and gelatin as substrates, one unit of collagenase activity was defined as the µmol of leucine equivalent per min/ml of the culture. For casein as substrate, one unit of collagenase activity was defined as the amount of enzyme that liberates 1.0 nmol of tyrosine s−1 under assay conditions. For azo-casein as substrate, a single activity unit was defined as the quantity of µmol of L-leucine liberated when exposed to 1 ml culture filtrate consisting collagenase, following 18 h at 37 °C (Wanderley et al., 2020) and for Hemoglobin as substrate, one unit of collagenase (protease) activity was defined as the amount of enzyme required to hydrolyze hemoglobin and release equivalent to 1.0 µg of tyrosine within 1 min reaction at 37 °C (Yin et al., 2013).
2.6.1.5 Determination of kinetic parameters
The kinetic parameters, which include the Michaelis – Menten constant (Km), maximal velocity (Vmax), turnover number (kcat), and kcat/km,Vmax/Km were calculated using the Lineweaver- Burk plot and equation based on the methodology described by Kerouaz et al., 2021; Teramura et al., 2011. The kinetic constants were evaluated by using collagen and gelatin as substrates at concentration from 0.4 to 2.0 mg/ml conducted under standard assay conditions.
2.7 Analytical characterization of recombinant collagenase
2.7.1 Mass spectrometry analysis
Tryptic enzymatic digests of collagenase expressed with MBP tag were subjected to mass spectrometry (MS) analysis in order to verify the authenticity of the recombinant collagenase. The digestion of collagenase-MBP protein was carried out by utilizing Trypsin-ultraTMmassspectrometry grade from New England Biolabs (NEB, Hertfordshire, UK). Trypsin stock solution (1 µg/µL) was produced by adding 20 µL of pure sterilized water to 20 µg lyophilized Trypsin- ultraTMand stored at −20 °C until further use. Trypsin digestion reaction for the recombinant collagenase protein was initiated with substrate (protein): trypsin ratio of 25:1. This included the addition of 1 µL of trypsin stock solution, 25 µL of protein, 2.5 µL of 2 × trypsin reaction buffer and 2.5 µL of ultra-purified water. The reaction mixture was kept for incubation in shaking condition for 16 h at 37 ± 2 °C and preserved at −80 °C. Reaction samples forMALDI-TOF-MS analysis was formed by desalting the reaction mixtures with C-18 Zip-Tip (Millipore). MALDI-TOF MS analysis was carried out at Mass Spectrometer facility, Department of Biological Sciences at the Indian Institute of Science (IISc), Bangalore using Sinapic acid as the matrix for analysis(Yaraguppi et al., 2022b).
2.7.2 2.7.2. N-terminal sequence identification by MALDI-TOF-MS and Edman degradation
In order to gain additional insight into N-terminal sequence of the peptide, confirm its authenticity, the digestion of recombinant collagenase was conducted as detailed in section 2.7.1. Subsequently, peptide mapping was executed using MALDI-TOF-MS. For the execution of mass spectrometric peptide mapping, a 0.5 µL of sample solution was spotted onto a stainless-steel target plate and permitted to dry. Further, 1 µL matrix was added on top of sample. The matrix solution consisted of concentrated solution of α-cyano-4-hydroxycinnamic acid (CHCA) dissolved in acetonitrile (ACN) and 0.1 % trifluoroacetic acid (TFA) in water. The sample and matrix solution were directly blended on target. The MALDI-TOF measurements was carried out employing a RapiflexX mass spectrometer (Bruker Daltonik, Bremen, Germany). At an accelerating voltage of 20 kV, the device was run in positive ion reflector mode with delayed extraction. For MALDI, a smart 3D laser beam (UV solid-state laser) with pulse width of 3–5 ns at 337 nm was employed (Meier-Credo et al., 2022; Duncan et al., 2016; Chotichayapong et al., 2016). To analyse peptides in reflector mode, the parameter settings were optimized. Sum spectra spanning from m/z from 500 to 4000 were produced by taking 1000 laser shots at different positions, depending on the signal-to-noise (S/N) ratio reflected on the oscilloscope. An external peptide calibration standard (Bruker Daltonik, Bremen, Germany) was used to calibrate the obtained spectra that is available commercially. Bruker flex control software RP_600-3200_Da.par method was employed to obtain sample data and further Bruker flex analysis v 4.2 program for used for data processing. Later Bruker Biotools software v 3.2, was used to carry out Protein sequence confirmation and N-terminal peptide sequencing (MTAVNQTISK with methionine oxidation) (Duncan et al., 2016; Torres-Sangiao et al., 2021).
The recombinant collagenase was subjected to Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel transfer onto the ProBlott membrane for subsequent N-terminal sequence analysis using automated Edman degradation via a protein sequencer (Shimadzu PPSQ-31A/33A) (Das et al., 2018; Laribi-Habchi et al., 2020). Identification of amino acid residues was accomplished through individual signal analysis. To perform an analysis of amino acid sequence similarity and multiple sequence alignments, the Protein BLASTwith the Swiss-Prot/TrEMBL(Translation of European Molecular Biology Laboratory) database was employed. Additionally, the Clustal Omega program, accessible on the European Bioinformatics Institute server (https://www.ebi.ac.uk/Tools/msa/clustalo/) was also utilized for these analyses. Dissimilar amino acids were uncovered. A comparison was performed between the N-terminal sequence of the recombinant collagenase and related collagenase sequences from Bacillus species found in the Swiss-Prot/TrEMBL database. This analysis aimed to ascertain identities in terms of percentage similarity (Laribi-Habchi et al., 2020).
2.7.3 CD spectroscopy analysis
The secondary structure of purified recombinant collagenase was examined in a liquid environment at 25 °C using a Jasco J-1500 Circular Dichroism Spectrophotometer, which was purged with N2 gas. The purified recombinant collagenase was diluted in 50 mM Tris HCl buffer (pH 7.0) resulting in final concentration of 50 µg/mL. Subsequently, CD spectra was recorded from 190 to 280 nm using a quartz cuvette with scanning rate of 50 nm/min. The CD spectra were recorded (CD mdeg) and transformed into molar ellipticity. To ensure accuracy, any influence from the solvent was eliminated by subtracting the CD spectrum of the solvent from that of the protein spectra. K2D2 software was used to determine the secondary structure and helical content of the protein (Indra et al., 2005).
2.7.4 1H NMR spectroscopy analysis
NMR spectroscopy (Jeol Resonance) operating at 400 MHz, was utilized to perform 1H NMR analysis on the purified recombinant collagenase. This analysis was conducted under room temperature (37 °C) conditions, with tetramethylsilane (internal standard). The purified collagenase was dissolved in Dimethyl sulfoxide (DMSO)as the solvent. Signals in the 1H NMR spectra were recorded within the range of 0–14 ppm, and the assignment of peaks in spectra was based on reported literature (Bagewadi et al., 2019).
2.7.5 TGA analysis
Assessment of thermostability of recombinant collagenase was done by TGA (Perkin-Elmer, USA). The sample (3 ml) was examined by heating at 25–800 °C) under nitrogen – controlled conditions with heating rate of 10 °C/min (Bagewadi et al., 2019; Bagewadi et al., 2020).
2.8 Application of recombinant collagenase in gelatin liquefaction
A gelatin liquefaction test was performed by inoculating purified recombinant collagenase into deep agar gelatin test tubes containing agar at concentrations of 1 % and 2 % and gelatin at 0.4 % and the test tubes were then incubated at 37 °C for 24 h. Isolates demonstrating the ability to induce liquefaction of the gelatin were considered as having potential applicability for the degradation of gelatin. This test serves as an indicator of the enzymatic activity of the recombinant collagenase and its capacity to hydrolyze gelatin substrates (Joseph et al., 2019).
2.9 In-silico studies
2.9.1 Structural and functional characterization of collagenase gene
The ExPASy(Expert Protein Analysis System)translate tool is employed for translating nucleic acid sequences (DNA/RNA) into corresponding protein sequences (amino acid sequences). The collagenase gene sequence of Bacillus sp.strain Z1 is submitted to ExPASy translate tool (https://web.expasy.org/translate/) online server to obtain translated protein sequence. The obtained amino acid composition of Bacillus sp.strain Z1 collagenase gene, was assessed for physical and chemical attributes utilizing the ExpasyProtParam online tool (https://web.expasy.org/protparam/). ProtParam is an in-silico tool utilized to compute diverse physiochemical parameters of proteins. The analysis involved evaluating amino acid composition, molecular weights, extinction co-efficient, theoretical isoelectric points (pI), aliphatic index, instability index, atomic compositions, estimated half-life, and the grand average of hydropathicity of selected protein(GRAVY) (Rani and Pooja, 2018). Collagenase protein sequence was submitted to EBI (European Bioinformatics Institute)- InterPro scan tool (https://www.ebi.ac.uk/interpro/search/sequence/) wherein the sequence is scanned to match related sequences and the tool offers a comprehensive categorization of protein sequences into groups, detecting important functional domains and key active sites. Phylogenetic trees were formulated by utilizing collagenase gene nucleotide sequence to explore the evolutionary connections among these genes. The gene sequences used in the study were from Bacillus siamensis, Bacillus velezensis and Bacillus amyloliquefaciensstrains. The full-length collagenase nucleotide sequences were retrieved from the NCBI database in FASTA format. The selected nucleotide sequences were aligned using multiple sequence alignment clustalW2 server. Phylogenetic tree of collagenase nucleotide sequence from different strains of Bacillussps. was analyzed using MEGA(Molecular Evolutionary Genetics Analysis) software. Similarly, phylogenetic tree of collagenase gene protein sequence was constructed with related collagenase gene protein sequences of Bacillus species. In this study collagenase gene protein sequences were retrieved from NCBI database in FASTA format. Total 5 protein sequences of collagenase gene from U32 family peptidase Bacillussps. with accession number namely Bacillus siamensis (WP_064777716.1), Bacillus siamensis (WP_19867931.1), Bacillus sp. JFL15 (WP_049627089.1), Bacillus amyloliquefaciens (WP_045510354.1) and Bacillus nakamurai (WP_061522884.1) were used in this study. Sequences of the selected protein was aligned using MSA clustalW2 server and using MEGA software phylogenetic tree was constructed. The evolutionary lineage of collagenase was done utilizing the neighbor-joining approach (Rani and Pooja, 2018).
Homology modelling is widely used technique to generate valid protein structure utilizing the amino acid sequence. To date, the crystal structure of collagenase gene from Bacillus siamensis is not established, so in the present study SWISS MODEL server (https://swissmodel.expasy.org/interactive) was employed for generating the homology model for collagenase gene from Bacillus siamensis. The protein sequence of collagenase gene was submitted to SWISS MODEL server, tRNA hydroxylation protein P1 (O32034.1. A) of Bacillus subtilis (strain 168) showed 93.13 % identity. After preparing the 3D structure of protein by SWISS MODEL server, the Protein Data Bank (PDB) file was downloaded and used for further analysis and molecular docking. The modeled structure stability and quality was checked by Ramachandran plot analysis by SWISS MODEL server, MOLPROBITY, ERRAT tools. The secondary structure of collagenase protein was included in MSA figure generation employing ESPript 3.0 (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The highly flexible regions of collagenase gene were predicted by bio-informatic tool FoldUnfold (https://bio.tools/foldunfold) (Su et al., 2022).
2.9.2 Molecular docking of collagenase with substrates
The molecular docking of homology modelled collagenase gene on four different substrates was carried out using PyRx software (version 0.8). The PyRx software is employed with Autodock Vina, Open Babel and Python Shell. The four substrates employed for molecular docking are APHCP (Alaska pollock hydroxyproline containing marine collagen peptide) (ID-129908191); Collagen type 4 alpha (531–543) (ID-16132108); PZ-peptide (ID-161208) and FALGPA (2-furanacryloyl-l-leucylglycyl-l-prolyl-l-alanine) (ID-6439513). SDF (Structured Data File) files of all substrates were downloaded from Pubchem server. Both the protein and substrates were loaded into PyRx software and collagenase protein was put as fixed. A grid box was designed to cover the whole protein, with an exhaustiveness parameter of 8 for all docking. The binding affinity of substrates was analyzed for inter-residue interaction. To validate the docking scores of complexes obtained from PyRx Autodocking Vina, the classic Autodocking was utilized. The protein and substrate interaction were generated using PyMOL (version 2.5.5) and Discovery studio visualizer (DSV) (version 21.1.0.0), from BIOVIA (Adeniji et al., 2020).
Molecular docking was also carried out using the Autodock software (autodock suite-4.2.6-×86_64 linux3.tar) as part of its execution process (Revankar et al., 2023). Autodock, an efficient web-based application, specializes in conducting computational screenings of ligands/substrates for specific target protein of interest. The modelled collagen protein is pre-processed by removing water molecules and added with hydrophilic hydrogens and Kollman charges and converted to pdbqt through the autodock software. The ligand is prepared by converting the.sdf files into.pdb by using pymol and converted to.pdbqt by employing autodock tools. Forming the grid box is a crucial step in the molecular docking process. A config file is prepared comprising x, y, z co-ordinates, energy range and exhaustiveness. Over here, the same x, y, z co-ordinates employed in PyRx software is used in autodock to known the binding stability of collagenase gene with 4 different substrates. Finally a command prompt is run to obtain output.pdbqt and log.pdbqt files. The outcomes from multiple docking are automatically assessed, resulting in the generation of a hierarchical list featuring the top 10 docked structures for Collagenase protein and from it the first docked structure with each substrate having highest docked score is considered for further hydrogen bond interaction analysis. The highest docking score pose achieved by the collagenase protein target with substrate is recognized as an optimal binding configuration and this outcome is selected for subsequent Molecular Dynamics and Simulations studies. Post-docking, the best scored docking pose of each substrate is visualized using Pymol tool to identify the interacting residues such as hydrogen bonds between the protein and substrates (Ligands).
2.9.3 Molecular dynamics and simulations (MDS)
In the study, modelled collagenase gene docked with APHCP using Autodock software is chosen for further Molecular dynamics (MD). GROningenMAchine for Chemical Simulations (GROMACS) software (2019.4) is used for molecular dynamics to determine the stability of substrate specificity between collagenase gene and APHCP. The study employed the Gromos54a7 force field combined with the single point charge (SPC) water model. A prodrug server was utilized to acquire Ligand topology. MD simulations were set at 300 K using Berendsen thermostat for 200 ns, maintaining pressure at 1.01325 bars similar to physiological conditions. The tool imports the complex for protein preparation, addressing any errors. Explicit solvents, Na + and Cl-ions were used for solvation and system neutralization by the steepest decent method. The system underwent complete energy minimization for the entire 200 ns of MD simulations. Two steps were used to equilibrate the system. Two phases were involved in the equilibration process, which involved a few thousand molecules and the collagenase – substrate complex. Using an NVT ensemble, wherein the number of simulated particles (N), simulation cell volume (V) and temperature (T) are all kept fixed throughout the simulation, first phase equilibration was carried out for 30 ps at 300 K. This stabilized the temperature of the system. The NPT ensemble herein Number of moles (N), Pressure (P) and temperature (T) will be conserved was carried out in second phase equilibration up to 30 ps NPT simulation per atom. In addition, the system was simulated using Berendsen at a time step of 5 ps while maintaining a constant temperature of 300 K and pressure of 1 atm. The input file name. coherence of grid maps, and presence of non-standard atoms were assessed to verify the accuracy of the MD simulations (Tohar et al., 2021; Yaraguppi et al., 2021; Yaraguppi et al., 2022a).
The Gromacs Package is employed for analyzing the data derived from Protein – substrate complex molecular dynamics. It computes the RMSD (Root Mean Square Deviation) value by evaluating the atoms within the collagenase protein matrix and those bound to the protein in the ligand. Throughout the Molecular dynamics simulation, various supplementary factors such as Rg (Radius of Gyration), SASA (Solvent accessible Surface Area), and the analysis of hydrogen bonds are considered. The tools gmx-rms (RMSD), gmx-rmsf (RMSF), gmx-gyrate (Rg), gmxhbond (hydrogen bond) and gmx-sasa (SASA) were used to calculate and examine these factors using commands present in Gromacs (Revankar et al., 2023). For plotting of data, the 200 ns trajectory is divided into segments of 20 ns each to facilitate analysis of plots compared to visualizing a continuous 200 ns stretch. Tools like PyMol and Visual Molecular Dynamics (VMD) are utilized for the analysis and visualization of the simulation – derived data. Additionally, the xmgrace tool is employed for generating graphs and plots derived from simulation.
2.9.4 Molecular Mechanics poison surface Area analysis (MMPBSA)
The MM-PBSA method assesses protein’s binding free energy (DG binding), employing the open-source tool g_mmpbsa to evaluate the interaction potential of drug molecules within the stable simulation structure. Specifically, the GROMACS utility g_mmbpsa (https://rashmikumari.github.io/g_mmpbsa/) is employed to perform the needed calculations (Revankar et al., 2023). MM-PBSA process is used with g_mmbpsa tool to assess various binding energy components. This assessment encompasses an analysis of the entropy contribution and energy engagement of individual amino acids, employing an energy deposition strategy. The final 50 ns of simulations, being stable, are used for DG computation in MMPBSA. The interaction affinity of the inhibitor with the unbound protein is derived from the difference between ligand-bound and free protein conformations, calculated by a specific formula as follows:
ΔG binding = G-complex – (G-protein + G – ligand) (Yaraguppi et al., 2022a).
2.10 Statistical analysis
All experiments and reactions were conducted in triplicates for each data point. The data is presented as mean ± standard deviation (SD). In the figures, the error bars depict the standard deviation and were computed using Microsoft Excel.
3 Results and discussions
3.1 Identification of gene of interest
A pivotal and foundational step in any recombination study involves the precise identification of target gene. The gene of interest is collagenase gene, and to identify this gene comprehensive bioinformatic analysis was conducted. The alignment of genetic sequences, encompassing both the collagenase gene and the amino acid sequence derived from various bacterial variants in relation tostrain Z1, was undertaken through the utilization of the CLUSTAL Omega Multiple Sequence Alignment Tool (Sievers et al., 2011). This sequence alignment process was conducted utilizing genetic data sourced from the expansive NCBI repository. The choice of the 422 amino acid coding collagenase gene, which spans a genetic length of 1269 base pairs, was predicated with 99 % genetic similarity to the collagenase gene present in strain Z1. Teramura et al., 2011, isolated collagenase gene from Grimontia (Vibrio) hollisae by cloning and sequencing method. Collagenase gene consisted of 2,301 nucleotides and amino acid sequencing comparison of 767 amino acids revealed high homology to several collagenases by employing CLUSTAL W2 program.
3.2 3.2. Cloning of collagenase gene into vector
Phenol chloroform extraction was employed to isolate genomic DNA that possessed a concentration of 1119 ng/µl. Custom primers were purposefully crafted utilizing the DNA sequence unique to the collagenase gene. These primers were instrumental in driving the amplification process of the collagenase gene through PCR. Amplification process plays a pivotal role in successful attainment of genetic recombination. In order to validate that the amplified product corresponded to target gene, electrophoresis was employed. The outcome of this electrophoretic analysis confirmed the presence of a solitary band, 1.27 kb and is depicted in Fig. 2. The amplified gene product from PCR was purified and inserted into pET28 (MBP Tagged) vector. This integration process was carefully facilitated through restriction digestion, employing the BamHΙ and NotΙrestriction enzymes. Subsequently, a ligation step was carried out, followed by the transformation of the construct into E. coli DH5α cells. Similarly to current research, ColA, the collagenase was obtained through the amplification process using genomic DNA extracted from B. cereus strain ATCC14579. Specifically, designed PCR primers was used to amplify ColA, lacking both the predicted signal peptide and propeptide regions. Subsequently, the PCR products flanked by BamHΙ and XhoΙ, was inserted into pGEX-6P-1 plasmid. This construct was then introduced into E. coli BL21, resulting in the creation of a fusion protein – GST-ColA as described by Abfalter et al., 2016.
The identification of positive transformed colonies was picked by antibiotic-based screening method. These selected colonies were cultured on agar plates infused with kanamycin. To validate the presence of collagenase target gene within these transformants, a series of screening steps was carried out. This included utilization of colony PCR amplification, and the outcomes were made visible through agarose gel electrophoresis. The colonies that exhibited positive transformation were isolated and their plasmids were extracted possessing a concentration of 365.9 ng/µL and were preserved for subsequent utilization. Similar to current approach, a recombinant serine alkaline protease from Bacillussp RAM53 was selected by antibiotic screening and grown in kanamycin plates and positive single colonies were examined by PCR colony test and transferred to E. coli BL21 expression host. Genetic engineering of these serine proteases makes them commercially industrially important organisms (Alei et al., 2023). A visual representation of the strategy employed for subcloning to construct the recombinant collagenase- pET plasmid can be found in the Fig. 1. In the present study, the collagenase gene was identified from Bacillus siamensis strain Z1 (accession number OR054215.1). The recombinant cloned sequence obtained from sequencing were subjected to alignment, and the alignment revealed a consistent correspondence with the reference sequences, thereby confirming the accuracy.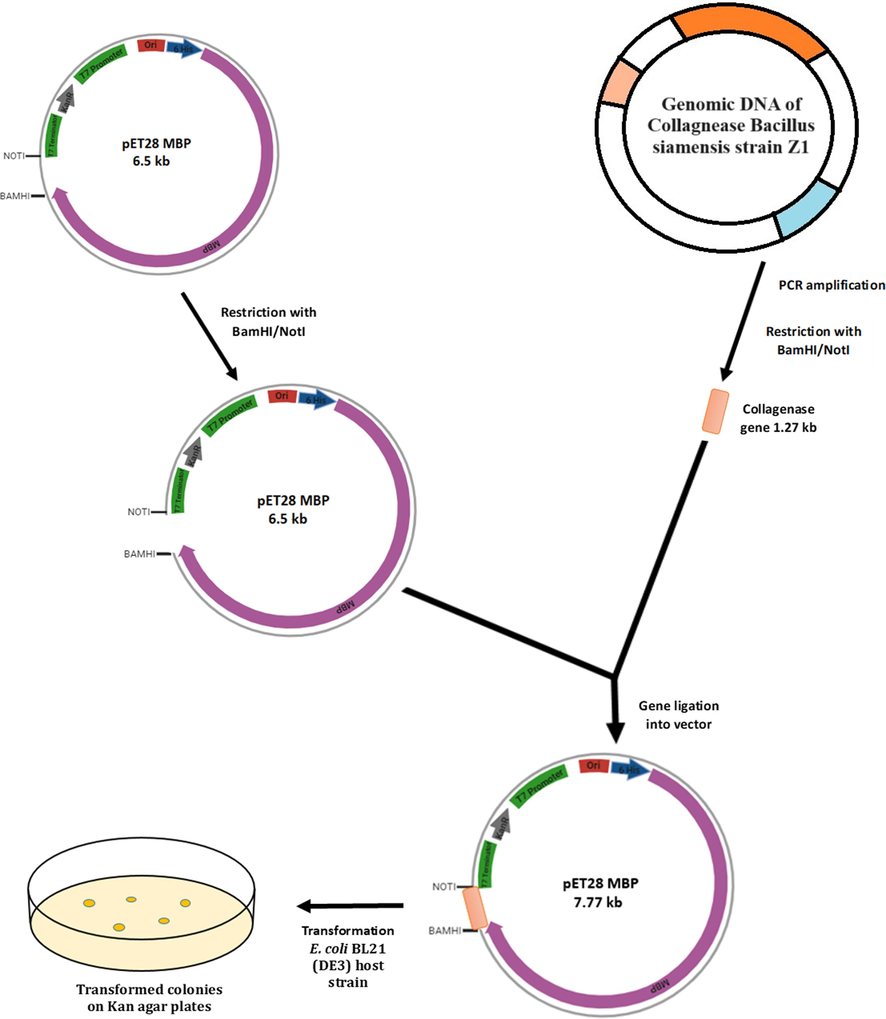
Illustrative representation of the subcloning process involved in constructing the recombinant collagenase.
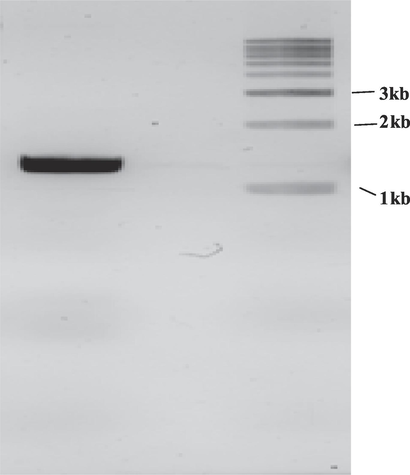
Amplification of collagenase gene by PCR from Bacillus siamensis strain Z1.
3.3 Expression and purification of recombinant collagenase
The genetic sequence was subjected to optimization and synthesis to ensure its adaptability for expression within the E. coli and collagenase gene of 1.269 bp was been cloned into pET28 (MBP Tagged) vector. The recombinant plasmids were transformed into E. coli DH5α cells. The plasmids extracted from these cells were transformed in E. coli BL21 (DE3) cells to enable their over expression. This over expression was carried out in sterile LB media supplemented with kanamycin (50 µg/ml) creating an environment that was favorable for the thriving of intended genetic process. The same procedure of expression with IPTG was also followed by Hu et al. (Hu et al., 2019) for expressing recombinant Matrix metalloproteinases (MMP1) HcMMP1. It is widely accepted and demonstrated fact that E. coli is renowned as one of the predominant microorganisms employed in the synthesis of recombinant proteins. Its widespread use can be attributed to several factors. E. coli has consistently been recognized for its adaptability as an expression system, characterized by its rapid growth, suitability for cultivation in cost effective culture media, and its ability to generate substantial cellular biomass while maintaining a rapid protein production rate (Alei et al., 2023). In present study expression of recombinant collagenase was induced by 1 mM IPTG. A concentration of 1 mM IPTG has been sufficient for induction of clostridial collagenases too (Eckhard et al., 2014). Ducka et al., 2009, reported usage of two concentrations (0.1, 1 mM) of IPTG for induction and cells were mainly induced by 0.1 mM and it was noted that no difference in soluble expression yield could be observed upon increasing IPTG concentration. The harvested cells were lysed and purified through Amylose resin column. The concentrated purified protein increased by 48.11 folds through buffer exchange. After 24 h of incubation, the crude recombinant protein from fermentation broth exhibited a collagenase activity of 117 U/ml and a protein concentration of 0.41 g/L. The MBP tag aided in one- step purification by amylose resin affinity chromatography, as detailed in Table 1. In comparison to cell lysate, collagenase obtained 20.76 folds increase in purity. The specific activity achieved of the purified collagenase was 5926.3 U/mg, which was significantly greater than the cell lysate. The chromatographic process yielded a recovery rate of 48.11 %. The purification of MBP fusion protein exploits the natural affinity of MBP for recombinant collagenase. It is important to view the MBP fusion tag as a means to enhance protein solubility and as an affinity tag for ease of purification process using column. Many vectors are available to construct fusion proteins with MBP (Duong-Ly and Gabelli, 2015). Amylose resin contains covalently linked amylose. Proteins tagged with MBP have a natural affinity for amylose and their specific binding allows for the isolation of target protein, while non-specific proteins and complex mixtures pass through (Pattenden and Thomas, 2008; Rodriguez et al., 2020). In accordance with this present research, recombinant L. sericata collagenase (MMP-1) expressed in SF9 cells showed crude activity to be 3.7 U/ml with 68.5 µg/ml concentration (Alipour et al., 2019). Zhu et al., 2022, reported purification of recombinant collagenase from B. subtilis WB600/pP43NMK-col was assisted by His – Tagged Nickel column resin yielding 2.14 % with 4.71 folds. Purification of collagenase from Bacillus pumilus Col-J was done yielding 7.0 % (Wu et al., 2010). Zhang et al., 2015 reported 73.2 % of relative activity recovery of recombinant collagenase with Histidine tagged (His-cdMMP-13).
Purification step
Total activity (units)a,b × 103
Total protein (mg)a,c
Specific acivity (U/mg of protein)
Activity yield (%)
Purification fold
Cell lysate
Amylose affinity chromatography58.5 ± 9.3
28.15 ± 1.3205 ± 16
4.75 ± 4.1285.36
5926.3100
48.111
20.76
The SDS-PAGE analysis of recombinant collagenase confirmed the successful expression of the gene, resulting in fusion protein with molecular weight of ∼ 89.4 kDa. This fusion comprised collagenase mass of 47.4 kDa and 42 kDa of MBP tag as observed in gel in Fig. 3. with protein molecular marker. It was found that the recombinant collagenase concentration was 203.356 µM. Fusion proteins have gathered interest owing to many advantages, including providing stability to the protein, augmenting protein solubility, and boosting folding characteristics. The signaling peptide is of crucial importance in the expression of fusion proteins, as it serves as recognition point for the secretory system. A larger concentration of released proteins was found in the culture broth because of the periplasm’s higher membrane permeability and easier downstream. The periplasm aids in protein folding, and the signal peptide facilitates the movement of recombinant proteins through the inner periplasmic membrane, enabling secretion in E. coli BL21 (DE3). This enhances soluble protein production (Shettar et al., 2023). Suberu et al., 2019, reported cloning of serine alkaline protease gene (R5) from Bacillus subtilis strain RD7 in pET15b and transformed in E. coli DH5α competent cells which is in line with this study. They expressed the gene in BL21 host cells and the product had a molecular weight of 43 kDa. Recombinant collagenase protein reveled a band of 52 kDa expressed of collagenase (MMP-1) (Alipour et al., 2019). An alkaline protease gene from Bacillus lehensis jo-26 was introduced into pET28 + a expression vector and transferred to E. coli BL21 expression host, yielding an enzyme with molecular weight of 34.6 kDa(Bhatt and Singh, 2020). Similarly, the serine alkaline protease gene from Bacillus sp. RAM53 was cloned into pET28a + vector, transferred to E. coli BL21, resulting in a recombinant enzyme with a molecular weight of 40 kDa (Alei et al., 2023). A recombinant keratinase was constructed using keratinase gene from Meiothermustaiwanensis WR-220 and expressed in E. coli expression system, further purified by affinity chromatography yielded molecular weight of ∼ 28.5 kDa. It also retained broad proteolytic activity for different substrates like feathers and casein and also retained activities in varying pH range 3 to 10 (Ho et al., 2016). In current study, the development of a highly effective approach for expressing and secreting recombinant collagenase into culture medium has led to a significant improvement in the recovery of product.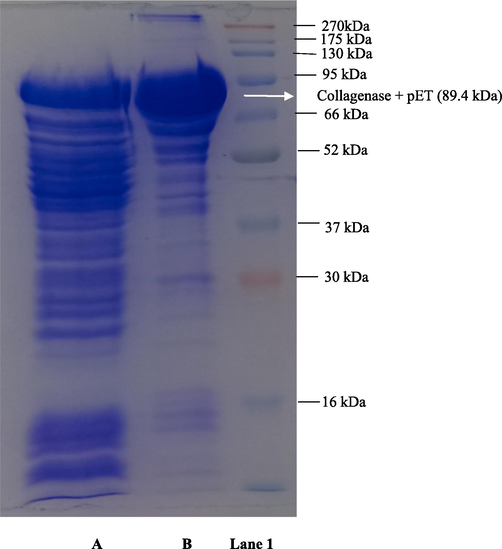
SDS-PAGE analysis of recombinant collagenase. (A) Z1 pET crude collagenase (B) Z1 pET MBP tagged Affinity purified collagenase (Lane 1) protein molecular weight marker.
3.4 Biochemical properties of purified recombinant collagenase
3.4.1 Effect of pH and temperature on the activity and stability of recombinant collagenase
The recombinant collagenase was highly active in alkaline pH range of 8––10 retaining > 90 % of the relative activity and with optimum pH at 9 and 10 as shown in Fig. 4A. In acidic pH range of 3–5, < 50 % of relative activity was observed. The reduction in the activity at acidic pH may be contributed by the variations occurring in the ionic forms of active sites of the enzyme further influencing the structure and folding attributes of proteins. However, at higher alkaline pH range of 11–12 the relative activities are evidenced to be 60 – 77 %. The pH stability of recombinant collagenase was studied by pre-incubating it at pH 9 and 10 for a period of 20 h at 35 °C. Collagenase showed a notable stability at pH 9 up to 20 h with > 95 % of relative activity as depicted in Fig. 4B indicating its alkaline nature. Fig. 4C illustrate the temperature effect on the collagenase activity. The optimum activity of the enzyme was observed at 50 °C at pH 9, and about > 75 % of the activity was retained between 40 and 60 °C. The enzyme activity declined sharply when the temperature exceeded 60 °C with less than 40 % of its activity retained. The recombinant collagenase possesses thermostability up to 50 °C. The thermostability was studied in the temperature range of 50 – 60 °C for 20 h. The half-life times of collagenase at 50, 55 and 60 °C were found to be 9, 7.5 and 5 respectively as shown in Fig. 4D. In context with present study, Zhu et al. (2022), reported recombinant collagenase from Bacillus subtilis WB600 showing maximum collagenase activity at pH 9.0. and it was relatively stable in the pH range 8–10 for 90 min by retaining ∼ 60 % of relative activity and optimum temperature was observed at 50 °C with retaining 75 % of its activity between 30 and 50 °C. A thermophilic collagenase from NocardiopsisdassonvilleiNRC2aza showed optimum activity at pH 8.0 and temperature of 60 °C, and stability at various pH levels (7.0–8.5) and temperatures (40–60 °C) (Abood et al., 2018). A recombinant serine protease by Bacillus sp. RAM53 displayed optimum activity at pH 9.0 and 40 °C temperature (Alei et al., 2023). A maximum collagenase activity of B. cereus isolated from pollen of bee of Amazon region was noted to be at 45 °C and pH 7.2 and had wide working ranges of pH values (7.2–10.0) and temperatures (25––50 °C) (Pequeno et al., 2019). The enzyme’s capacity to withstand high alkalinity is a crucial attribute for its utilization across various industries. Notably, the recombinant collagenase displayed high alkalinity and slight thermostability, the inclusion of MBP tag might also play significant role in enhancing the enzyme’s stability.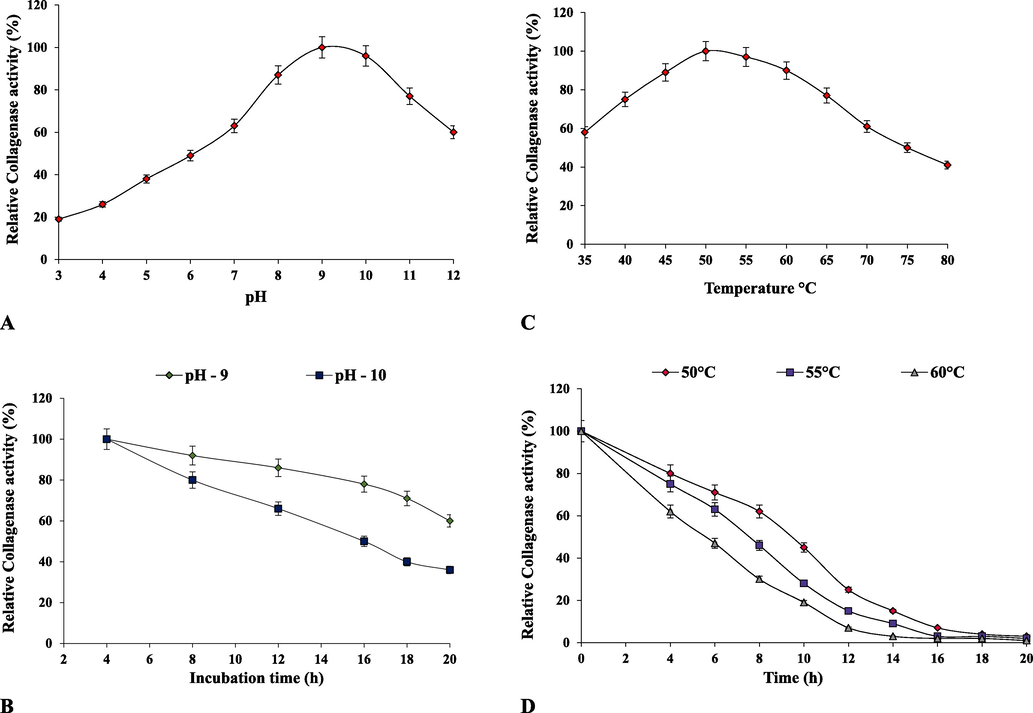
Biochemical characterization of purified recombinant collagenase. (A) effect of pH on collagenase activity, (B) pH stability, (C) effect of temperature on collagenase activity and (D) temperature stability.
3.4.2 Effect of metal ions, chemicals, organic solvents and additives
The effect of some metal ions such as FeSO4, MgSO4, NaCl, KCl, CaCl2, ZnSO4, CuSO4 and MnSO4 on the enzyme activity was evaluated (Table 2). The catalytic efficiency of the recombinant collagenase was stimulated by CaCl2, MgSO4 and FeSO4and MnSO4 with > 85 % activity retained. The biggest stimulation was observed in the presence of CaCl2with 110 % relative activity, while the notable inhibition was observed in presence of KCl with relative activity of 41 %. A similar collagenase relative activity enhancement was observed with metal ion Ca2+ on rBLAP recombinant collagenase (Joshi and Satyanarayana, 2013). Li et al., 2022, reported novel marine gelatinase Flocculibactercollagenilyticus also possessing collagenase activity. Metal ions like Ca2+, Ba2+, Mg2+ significantly increased activity towards bovine bone collagen, but inhibited by Zn2+. Metal ions such as Zn2+, Co2+, Mg2+ and Ca2+ stimulated the recombinant collagenase activity, but was inhibited by Fe3+ and Cu2+(Zhu et al., 2022), and in another study collagenase activity was inhibited by Na+ and K+, while stimulated by Ca2+, Mg2+ and Fe2+(Al-Bedak et al., 2022). The relative collagenase activity in absence of any surfactants, organic solvents, metal ions and additives was considered as control.
Factors
Relative Collagenase activity (%) a
Control
Metal Ions
Surfactants
Organic Solvents
AdditivesWithout treatment
FeSO4
MgSO4
NaCl
KCl
CaCl2
ZnSO4
CuSO4
MnSO4
SDS
Triton X-100
Tween 80
Methanol
Ethyl acetate
Benzene
Chloroform
Urea
LD-DTT
β – mercaptoethanol
EDTA100
88
102
50
41
110
78
80
85
94
96
92
60
55
52
43
88
93
96
72
The influence of surfactants such as SDS, Tween 80 and Triton X-100 on the collagenase relative activity is illustrated in Table 2. All the surfactants have shown > 90 % of relative activity. Corresponding to present study, an ionic detergent SDS has shown stimulatory activity on recombinant rBLAP collagenase assuming the SDS must have assisted in attaining favorable conformation of protein rather than the amphiphilic nature of SDS, that allows interactions with amino acids leading to unfolding of protein and loss of enzyme activity (Joshi and Satyanarayana, 2013). Wanderley et al. (2020), reported collagenase activity from Chlorella vulgaris for surfactants such as Triton X, Tween 80 and SDS to be 40 %, 74 % and 62 % respectively. Free collagenase and collagenase nanoparticles of Bacillussp when treated with same surfactants exhibited stability (Badoei-dalfard et al., 2022). Recombinant serine protease displayed increase in enzyme activities in presence of surfactants, SDS is a serine protease activator and indicates the suitability of SDS as detergent and is suggested that positive charges of calcium and magnesium found in hard water combine with negative charges of SDS to deactivate them (da Silva et al., 2018).
It can be seen that organic solvents like Methanol and ethyl acetate have retained > 50 % relative activity as there is a high demand for alkaline proteases that maintain stability in organic solvents because they can efficiently catalyze product synthesis in organic environments, while aromatic solvents such as benzene and chloroform have displayed slight inhibitory activity (Table 2). Inhibitory activity of recombinant collagenase was observed by aromatic solvents such as benzene and toluene and this enzymatic catalysis changes might be due to factors such as the disruption of hydrogen bonds, alterations in hydrophobic interactions, enzyme compaction and modifications in protein conformation (enzyme unfolding) and inactivation. A study exhibited that aromatic solvent benzene adversely affected collagenase activity, whereas methanol enhanced the activity (Joshi and Satyanarayana, 2013). Similar result of inhibition was displayed by free collagenase (Badoei-dalfard et al., 2022) and activity enhancement by methanol (Taghizadeh Andevari et al., 2019).
The stability of collagenase was assessed by treatment with inhibitors. Among the inhibitors tested, EDTA, a well-known metalloprotease inhibitor inhibited the collagenase thus indicating recombinant collagenase gene is a metalloprotease. LD-DTT and β – mercaptoethanol had a slight enhancing activity on collagenase, indicating the involvement of thiol groups in proteolysis (Table. 2). In accordance with current results EDTA had shown inhibitory activity, whereas β – mercaptoethanol and LD-DTT showed slight stimulatory activity on recombinant collagenase rBLAP (Joshi and Satyanarayana, 2013). Similar activity reducing profile was displayed by EDTA on collagenolytic metalloprotease isolated from banana peel and stated higher concentration inhibition by metalloprotease inhibitor classifies enzyme as metalloprotease (Gurumallesh et al., 2019). EDTA acts as chelating agent that binds and removes metal ions from active metalloproteases. Consequently, this action results in inactivation of metalloprotease (Ghauri et al., 2022; Laustsen et al., 2016).
3.4.3 Thermodynamic analysis of collagenase
The thermodynamics analysis of recombinant collagenase is essential for understanding stability and behavior under varying temperature conditions. Table 3. presents thermodynamic attributes of recombinant collagenase. Rise in temperature from 50 to 60 °C showed decrease in t1/2 and D values. A notable rise in thermal stability was observed at 70 °C. An apparent correlation was seen between the increase in temperature and increase of deactivation rate constant (Kd). The enthalpy of denaturation (ΔHd) quantifies the amount of heat absorbed during the denaturation of protein. A greater ΔHd signifies increased enzyme thermostability. The ΔHd values appeared to decrease as temperature rised, suggesting minimal energy needed for enzyme denaturation. This might relate to structural alterations in proteins at elevated temperatures. Gibbs free energy (ΔGd) combines enthalpy and entropy, and a robust tool for precise enzyme stability assessment. Higher temperatures leading to increased ΔGd values suggest a direct correlation between protein stability and elevated ΔGdvalues, indicating a non-spontaneous thermal inactivation mechanism. The entropy ΔSd is enzyme thermal denaturation, representing the energy/degree in the shift from the native to denatured state. Increased ΔSd signifies increased disorderness linked to enzyme structure, observed with elevated temperatures. Negative ΔSd valued denote non-spontaneous reactions and also highlight enzyme is stable during thermal denaturation process. The thermodynamic factors notably indicate the thermostability of recombinant collagenase. The thermodynamic characteristics of the enzymes associated with thermal deactivation are also been addressed by other researchers as well. Costa et al. (2023), have showed thermodynamic properties with ΔGdand ΔSd value of 62.16 kJ/mol and 1.96 J/mol respectively of collagenase from Streptomyces antibioticus. A lower t1/2 and Kd values in range of 0.1 – 2.0 h and 0.0035–––0.057 m-1 were observed for purified alkaline protease from N. dassonvillei OK-18 at varying conditions (Sharma et al., 2020). Thermal inactivation of free and immobilized alkaline serine protease showed extended half-life and greater thermal stability in bead form. Higher values of kinetic parameters like enthalpy denaturation, ΔGd, entropy was observed in immobilized enzyme (Mechri et al., 2022). A serine protease thermodynamic attributes such as Kd and t1/2 ranged from 2.50 × 103to 5.50 × 103and 277.25––111.25 min respectively within temperature range of 50 – 80 °C(Chauhan et al., 2021).
Temperature (°C)
Half-life t1/2 (h)
Kd (h−1)
D (h)
ΔHd (kJ.mol−1)
ΔGd (kJ.mol−1)
ΔSd (kJ.mol−1)
50
55
609
7.5
50.11
0.13
0.220.93
17.71
11.5116.74
16.63
16.5185.12
85.26
85.79−0.211
−0.209
−0.207
3.4.4 Evaluation of substrate specificity
To assess the substrate specificity of recombinant collagenase, the relative activities (%) were examined using various types of substrates as detailed in Table 4. Recombinant collagenase showed the hydrolysis of natural substrates such as collagen, gelatin, BSA, casein and hemoglobin and azo-casein (modified). Collagenase demonstrated outstanding catalytic performance when acted on collagen and gelatin with > 100 % relative activity. The collagenase also acted on other natural and modified substrates with retaining > 90 % relative activity and 40 % of activity was observed with hemoglobin. Badoei-dalfard et al., 2022, showed higher affinity of free and immobilized collagenase nanoparticles towards collagen about 2.44, 4.05 folds time compared to casein. It also showed activity towards fibrin acting as fibrinolytic protease in hydrolysis of blood clots. The present study finding aligned with prior research demonstrating significant substrate specificity towards bovine bone collagen and gelatin, as illustrated involving recombinant gelatinase Aa2_1884 (Li et al., 2022). In accordance to present results, Higher affinity of gelatin and collagen towards ATPS purified collagenase produced by microalgae C. vulgaris was reported by Wanderley et al. (2020). Proteolytic activity towards other substrates such as casein, azocasein, BSA. and hemoglobin were reported by other researchers (Suberu et al., 2019; Yin et al., 2013; Sriket et al., 2011).
Substrate
Concentration
Relative Collagenase activity (%) a
Collagen
Gelatin
BSA
Casein
Azo-casein
Haemoglobin10 g/L
2 g/L
10 g/L
20 g/L
30 g/L
20 g/L100 ± 2.3
115 ± 1.4
98 ± 0.8
92 ± 1.5
90 ± 2.2
43 ± 1.9
3.4.5 Determination of kinetic parameters
The Table 5 showcases the analyzed kinetic parameters of purified recombinant collagenase, illustrating its adherence to the classical Michaelis-Menten kinetics, as revealed through the Lineweaver-Burk plot analysis. The activity of Collagenase progressively elevated up with increased substrate concentration and reached saturation point, signifying the complete occupancy of active sites. The Km values for collagen and gelatin was found to be 0.33 mg/ml and 0.43 mg/ml respectively. Vmax values were found to be 5.926 × 103 and 6.432 × 103 U/mg respectively. Lower Km value indicates increased substrate specificity and strong substrate binding. The observed Km values signify the increased affinity of the gelatin substrate for collagenase. A lower Km and Vmax of 1.09 × 10-3 and 4.06 × 10-4 was observed for recombinant collagenase showing good affinity and catalytic efficiency for collagen (Tanaka et al., 2020). Vmax refers to the maximum rate achieved when the enzyme is fully saturated with substrate. The calculated Kcat and deduced catalytic efficiency Kcat/Km for collagen was 11.852 × 103 min−1 and 35.915 × 103 min−1 mM−1 respectively and for gelatin was 12.864 × 103 min−1 and 29.916 × 103 min−1 mM−1 respectively. Specific constant (Vmax/Km) for collagen and gelatin were found to be 17. 95 × 103 and 14.95 × 103respectively. A Km value of 3.03 µg/ml and Vmax of 0.15 µg/ml min−1 was observed for EDTA inhibited gelatinase towards gelatin (Silvestre and Ramos, 2015). Kinetic parameters were studied for collagenase produced with Kcat/Km value of 2.95 mL/mg/s (Costa et al., 2023). A mutant keratinases derived from Brevibacillusparabrevis hadKcat/Km varying in range 11 to 20 s−1 mM−1, while the wild exhibited 12.5 s−1 mM−1(Shi et al., 2021). Abdella et al., (2023), demonstrated enhanced Km and Vmax in the immobilized enzyme, suggesting improved enzyme activity post-immobilization, due to enhanced substrate conversion rates. The elevated Vmax might be from enzyme-carrier binding, enhancing active site affinity for substrate interaction.
Substrate
Km (mg/ml)a
Vmax (×103 U/mg) a
kcat (×103 min−1)
Kcat/Km (×103 min−1 mg−1)
Vmax/Km (×103 min−1/mg−1)
Collagen
Gelatin0.33
0.435.926
6.43211.852
12.86435.915
29.91617.95
14.95
3.5 Analytical characterization of collagenase
3.5.1 Mass spectrometry analysis
The collagenase protein with MBP tag was subjected to enzymatic digestion using trypsin, an enzyme known for cleaving at the C-terminal of arginine (R) and lysine (K). The digestion was carried out with collagenase protein to trypsin ratio 25:1 and the resulting fragments were subsequently analysed using MALDI-TOF-MS. Table 6. displays the mass profiles of predicted and observed fragments resulting from the trypsin digestion of collagenase protein. The predicted mass fragments were calculated based on the primary protein sequence of collagenase by employing the EXPASY: PeptideMass tool (https://web.expasy.org/peptide_mass/). The mass spectra unveiled a total of 21 [M+H]+ fragments for collagenase – MBP with 68.7 % sequence coverage. Therefore, the data obtained from MALDI-TOF-MS analysis for the digestion of recombinant collagenase validates authenticity of the collagenase protein. Analogous analyses are reported in diverse literatures. Using MALDI-TOF-MS/MS analysis, the functional collagenase of Pseudoalteromonasagarivorans NW4237 was recognised as a member of U32 family peptidases, exhibiting a molecular mass of 52,509 Da (Bhattacharya et al., 2019). In a study molecular weight of collagen hydrolysates (peptides) produced from digestion by Penicillium aurantiogriseum URM 4622 were analysed by MALDI-TOF-MS ranging between 11 – 2 kDa (Lima et al., 2015). Saxena and Singh, 2015, analysed the masses of peptide fragments obtained by MALDI-TOF MS were in range 600 – 3000 Da produced by fibrinolytic collagenase. Thakrar et al., 2023, proposed recombinant alkaline serine protease gene from haloalkaliphilic actinobacteria, Nocardiopsis sp. had a molecular weight of 34 kDa determined using Mass spectrometer (MALDI-TOF). Liu et al., 2011, reported a molecular mass of 5200 and 5206 Da for two bacteriocins, enterocin 7A and 7B respectively, produced by Enterococcus faecalis 710C.
Predicted masses of collagenase,
[M+H]+
Observed masses of collagenase-MBP,
[M+H]+
2614.2493
2282.0943
2225.0729
2152.1033
2136.0872
2069.9661
1917.0071
1914.9252
1824.8836
1881.9509
1682.9509
1379.7892
1188.5612
1082.6357
1047.4993
1011.4750
984.3444
778.4246
718.3366
520.2296
175.11892614.917
2282.929
2225.930
2152.851
2136.825
2069.761
1917.732
1914.671
1824.733
1882.697
1682.779
1379.803
1189.617
1082.513
1047.369
1011.531
983.437
778.303
718.186
520.136
175.754
3.5.2 N − terminal sequence analysis by MALDI – TOF – MS and Edman degradation
In order to understand the functional characteristics of peptides and proteins, it is vital to determine their structure and identity. Edman sequencing, a conventional approach, offers high sensitivity in identifying the structural details of bioactive peptides. However, with the advancements in analytical methods, advanced mass spectrometry emerges as an alternate technique for the identification and mapping of peptides. Amino acid sequencing analysis can be performed utilizing MALDI-TOF-MS, liquid chromatography – tandem mass spectrometry (LC-MS) or quadrupole time-of-flight mass spectrometry (Q-TOF-MS). MALDI-TOF-MS was utilized for conducting the N-terminal sequence analysis of the recombinant collagenase. The resulting spectra was used to identify matching proteins in the NCBI database (https://www.ncbi.nlm.nih.gov/) using the Mascot search program (https://www.matrixscience.com, Matrix Science, London, England) or using online tools to confirm sequence coverage of known sequences. There was a mass range of 500–––4500 m/z. The mass spectra showed 21 fragments of [M+H]+ collagenase – MBP (Table 6). Additional analysis of tryptic peptides from 47.4 kDa protein was conducted to obtain sequence information from peptide ions, to confirm the identity of collagenase. Using parameters like fragment peak width of 0.2 m/z and signal-to-noise threshold of 2, the peaks were labelled using flex analysis software. Techniques including de-isotoping and centroiding of the spectrum were utilized. These techniques were employed to encompass the monoisotopic masses in peptide mass fingerprinting (PMF). This highlights the importance of identifying the monoisotopic masses in MALDI-TOF-MS for the structural analysis of the collagenase protein. Biotools software version 3.2 was used to analyze and validate the N-terminal sequence of the collagenase protein by applying it to the MALDI-TOF spectra. Parameters for sequence confirmation was set, including enzyme: trypsin, ion mode: positive, optimal modifications of methionine oxidation, and subsequent adjustments specifying methionine at position 1 and oxidation modification. The trypsin digestion uncovered a sequence starting with 10 amino acid residues at the N-terminus, identified as MTAVNQTISK, presenting a mass of 1092.57 m/z (calculated) and 1092.381 m/z (measured) via Biotools software v 3.2, as indicated in Table 7. Fig. 5 A1, A2 illustrates collagenase peptide fragmentation. Here, 35 peaks including the confirmation of the N-terminal peptide sequence are labelled according to their individual masses in m/z. The complete protein sequence is been provided in Supplementary Information (SI-S1) with matching sequences having an MS coverage of 63.8 %. Since trypsin cleaves specifically at R (arginine) and K (lysine) amino acids, it specifically cleaved at the K amino acid at position 10, confirming the peptide’s amino-terminal nature. Similar methodologies have been adopted by other researchers in their respective studies. The Edman degradation process unveiled 27 aa residues at the N-terminal sequence of the serine alkaline protease SAPGB from Gracilibacillusboraciitolerans LO15, showing the closest identity to the S8 protease family (Ouelhadj et al., 2020). A protein of bovine neutrophil gelatinase associated lipocalin (bNGAL) with relative molecular mass of ∼ 25,000 Da showed N-terminal homolog with hNGAL, 24p3 and A2UMRP having 68, 54 and 49 % of identity and 7 significant peaks were observed by MALDI-TOF Analysis (van Veen et al., 2006). The identity of a recombinant collagenase that is highly expressed is clearly shown by the current analysis.
Meas. m/z
Calc. MH+
Range
Sequence
1092.381
1114.49
745.269
1379.642
2069.758
2629.905
2294.904
1739.8
2152.847
778.302
718.199
557.238
1802.675
876.317
1522.548
1537.568
2272.94
2288.88
1932.718
2678.941
2224.925
1897.655
1753.799
1493.575
1882.702
1047.36
520.135
2544.84
2776.062
555.166
1082.509
1552.592
1267.527
1011.529
1043.359
1139.428
1324.5151092.57
1113.71
744.509
1379.789
2069.966
2630.244
2295.274
1739.972
2152.103
778.425
718.337
557.377
1803.004
876.467
1521.78
1537.778
2273.213
2289.208
1933.002
2679.377
2224.191
1897.04
1752.95
1493.81
1881.91
1047.5
520.23
2545.1
2775.37
555.293
1082.636
1552.921
1267.665
1011.475
1043.465
1139.57
1324.69
1–10
11–20
16–21
21–33
56–74
80–101
102–123
108 – 123
125––143
144 – 149
150 – 155
156 – 160
156 – 171
161 – 168
161 – 173
161 – 173
245 – 264
245 – 264
248 – 264
248 – 270
265 – 283
269 – 284
269 – 283
271 – 283
285 – 300
301 – 308
309 – 312
313 – 335
337 – 360
361 – 364
399 – 407
399 – 411
411 – 420
412 – 419
412 – 419
412 – 420
412 – 422MTAVNQTISK
VVNGKRVITK
RVITKK
KPELLAPAGNLEK
SNADNFSIEEIAEGVEFAK
IYVTTNIFAHNENMDGLEEYLK 14: Oxidation (M)
ALGDAKVAGIIVADPLIIETCR 21: Carbamidomethyl (C)
VAGIIVADPLIIETCR 15: Carbamidomethyl (C)
VAPDVEIHLSTQQSLSNWK
AVQFWK
EEGLDR
VVLAR
VVLARETSGLEIKEMK
ETSGLEIK
ETSGLEIKEMKEK
ETSGLEIKEMKEK 10: Oxidation (M)
DLKLIESIPQMIEMGIDSLK
DLKLIESIPQMIEMGIDSLK 14: Oxidation (M)
LIESIPQMIEMGIDSLK 8: Oxidation (M)
LIESIPQMIEMGIDSLKIEGRMK 8: Oxidation (M) 11: Oxidation (M) 22: Oxidation (M)
IEGRMKSIHYVATVVSVYR 5: Oxidation (M)
MKSIHYVATVVSVYRK 1: Oxidation (M)
MKSIHYVATVVSVYR
SIHYVATVVSVYR
VIDAYCADPENFVIQK 6: Carbamidomethyl (C)
EWLDELDK
CANR 1: Carbamidomethyl (C)
DTAPAFFEGTPGFEEQMFGEHGK 17: Oxidation (M)
TTFDFAGLVLAYNEETQMVTLQQR
NFFK
HPLQIVTFK
HPLQIVTFKVDKK
KIYPSNMMRK
IYPSNMMR
IYPSNMMR 6: Oxidation (M) 7: Oxidation (M)
IYPSNMMRK
IYPSNMMRKGK
The Edman degradation method used for N-terminal sequencing of the blotted recombinant collagenase (30 aa) revealed the sequence as, affirming the consistency and purity of the purified enzyme. Using the GenBank non-reductant protein database, the amino acid sequence was compared to the available protein sequences using the BLASTP search program. The sequence displayed slight varying degrees of similarity, showing the highest homology within the Bacillus species family.
Table 8 revealed the sequence similarity to U32 family peptidase (WP_064777716.1) from Bacillus siamensiswith 99.76 % similarity, U32 family peptidase (WP_198697931.1) from Bacillus siamensiswith 99.76 % identity, U32 family peptidase (WP_049627089.1) from Bacillus sp. JFL15 with 99.76 % identity, U32 family peptidase (WP_045510354.1) from Bacillus amyloliquefacienswith 99.29 % identity, U32 family peptidase (WP_061522884.1) from Bacillus nakamuraiwith 98.10 %, U32 family peptidase (WP_059335499.1) from Bacillus halotoleranswith 93.13 % and U32 family peptidase (MCY7851673.1) from Bacillus spizizeniiwith92.89 % identity. The high similarity of the amino acid sequences with related collagenases shows that recombinant collagenase is a distinct addition to the U32 family peptidase that falls within the classification microbial collagenases.
Enzyme
Origin
N – terminal amino acida,b
Identity (%)
Recombinant Collagenase (Current study)
U32 family peptidase (WP_064777716.1)
U32 family peptidase (WP_198697931.1)
U32 family peptidase (WP_049627089.1)
U32 family peptidase (WP_045510354.1)
U32 family peptidase (WP_061522884.1)
U32 family peptidase (WP_059335499.1)
U32 family peptidase (MCY7851673.1)
Bacillus siamensis
Bacillus siamensis
Bacillus siamensis
Bacillus sp. JFL15
Bacillus amyloliquefaciens
Bacillus nakamurai
Bacillus halotolerans
Bacillus spizizenii
MTAVNQTISKVVNGKRVITKKPELLAPAGN
MTAVNQTISKVVNGKRVITKKPELLAPAGN
MTAVNQTISKVVNGKRVITKKPELLAPAGN
MTAVNQTISKVVNGKRVITKKPELLAPAGN
MTAVNQTISKVVNGKRVITKKPELLAPAGN
MTAVNQTISKVVNGKRVITKKPELLAPAGN
MTAVNDKISKIVDGKRVITKKPELLAPAGN
MTAVNDKISTIVNGKRVITKKPELLAPAGN−
99.76 %
99.76 %
99.76 %
99.29 %
98.10 %
93.13 %
92.89 %
In Fig. 5B, the alignment comparison of the obtained recombinant collagenase (sequence) specifically with U32 family peptidases with accession number WP_045510354.1, WP_049627089.1, WP_198697931.1 and WP_064777716.1 shares the amino acid residue identities of 419/422, 421/422, 421/422, 421/422 respectively. Moreover, the highest sequence identity was obtained with U32 family peptidase Bacillus siamensis (WP_045510354.1) (99.76 %) with 3 aa differing namely, D233G, E235D and D307E and when compared to all selected Bacillus strains, 6 aa differed namely, D128E, D217D, D233D, E235E, V275V, D307D. In correlation to this research a serine protease EuRP-61 from Euphorbia resinifera latex had a molecular weight of 61 kDa analyzed by MALDI-TOF-MS and N-terminal sequence of 15 amino acids having 50–70 % of homology with other plant serine protease was identified by Edman degradation (Siritapetawee et al., 2020). Asmani et al., 2020, reported N-terminal sequence of first 27 amino acids of acido-thermostable purified chitinase (ChiA-Ba43) displaying homology to chitinases from other Bacillus species with 70 – 85 % of sequence identity and a molecular mass of 43,190.05 Da was determined by MALDI-TOF-MS. ChiA-Ba43 possessed alanine as first aa residue implying that a signal peptide responsible for secretion is cut from precursor to form mature extracellular ChiA-Ba43. N-terminal sequencing of 27 aa of Chitinase (ChiA-Si40) was analyzed by Edman degradation by Laribi-Habchi et al., 2020. The sequencing displayed variability of 66–––68 % identity with chitinases produced by Aeromnas sp. and S. piezotolerans and molecular weight of 40 kDa was determined by MALDI-TOF-MS.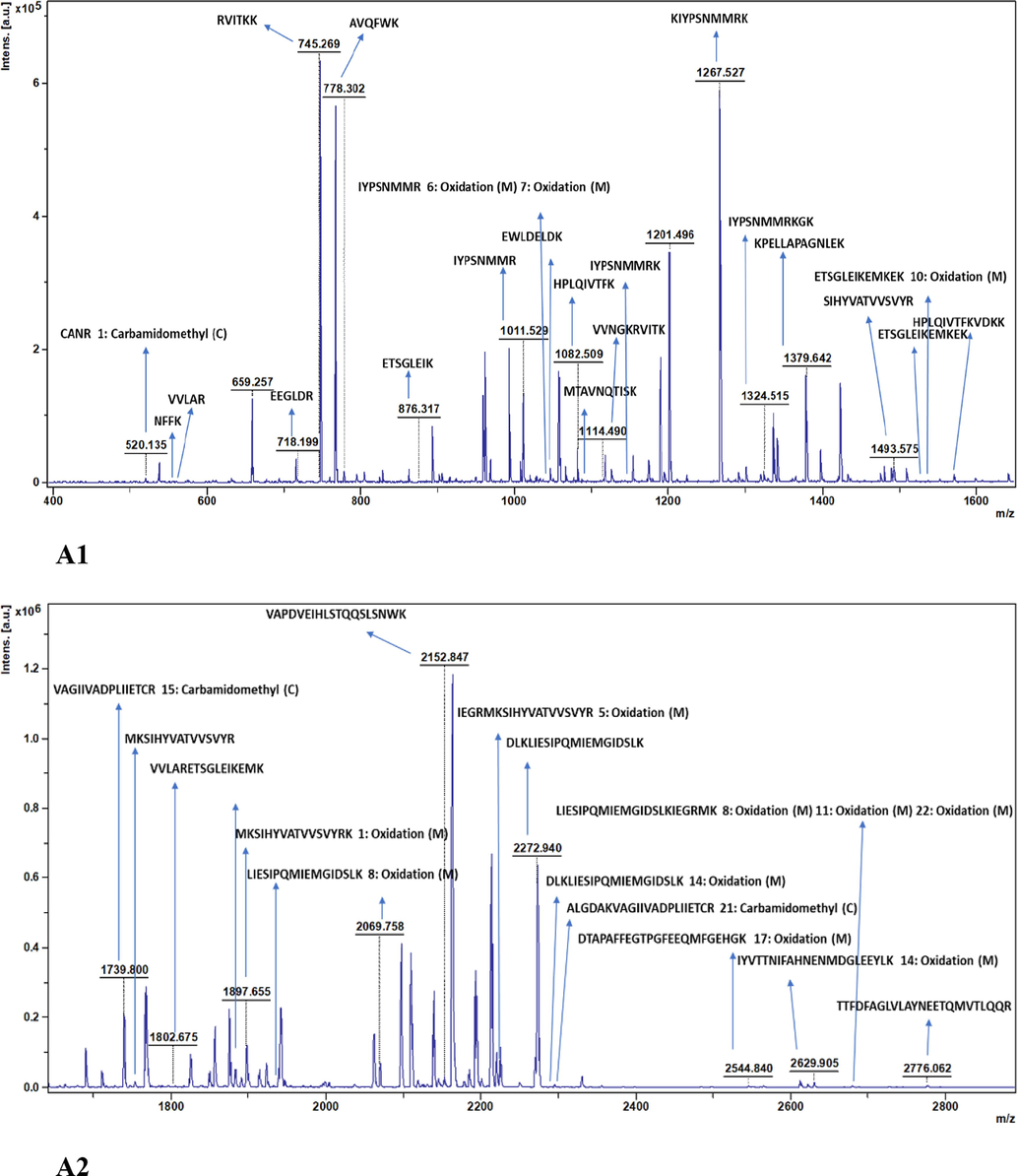
Analysis ofN −terminal sequence of recombinant collagenase. (A) MALDI-TOF spectra used for determination of protein sequence using biotools software v 3.2.(B) Multiple sequence alignment of recombinant Collagenase gene sequence with collagenases of U32 family WP_045510354.1, WP_049627089.1, WP_198697931.1 and WP_064777716.1. Numbers written above the lines indicate positions of amino acids. Residues that are identical are denoted in red boxes. Six different residues in the recombinant collagenase sequence compared to U32 family Bacillussps are marked with maroon stars. Symbols such as “α”, “β” and “T” represent α – helices, β- strands and β-turns respectively.
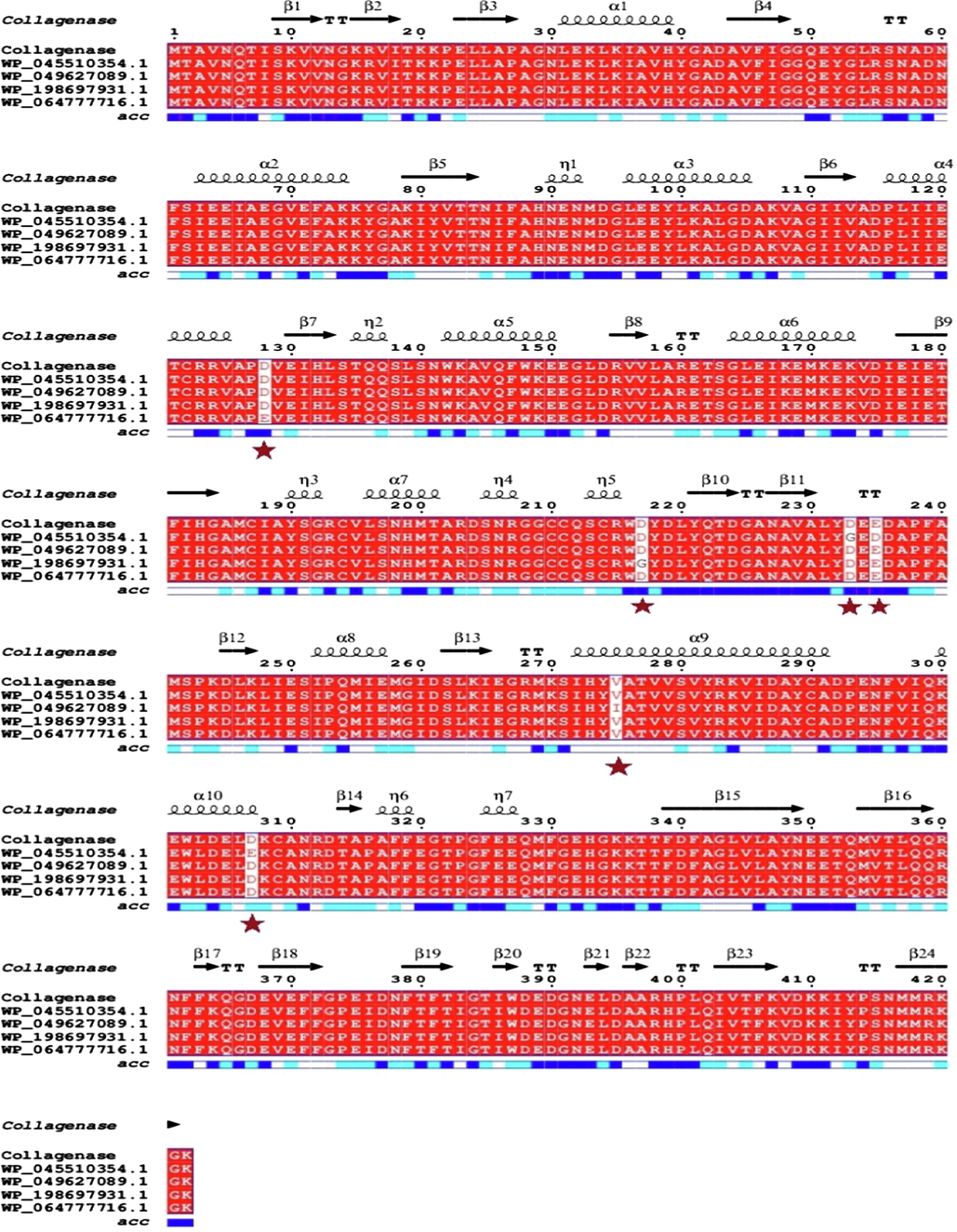
Analysis ofN −terminal sequence of recombinant collagenase. (A) MALDI-TOF spectra used for determination of protein sequence using biotools software v 3.2.(B) Multiple sequence alignment of recombinant Collagenase gene sequence with collagenases of U32 family WP_045510354.1, WP_049627089.1, WP_198697931.1 and WP_064777716.1. Numbers written above the lines indicate positions of amino acids. Residues that are identical are denoted in red boxes. Six different residues in the recombinant collagenase sequence compared to U32 family Bacillussps are marked with maroon stars. Symbols such as “α”, “β” and “T” represent α – helices, β- strands and β-turns respectively.
MALDI-TOF-MS, an analytical method, is recognized for its minimal sample destruction. Its high sensitivity necessitates only a small portion of the protein digest for mass analysis allowing the remaining portion to be available for additional measurements. Simply measuring the mass of a peptide is insufficient for obtaining its amino acid sequence; inducing peptide fragmentation is also crucial for acquiring this information. PMF involves analyzing the mass-to-charge ratio (m/z) of digested proteins and employing different online tools or search engines for identification. In this study, the acquired MALDI-TOF spectra were additionally utilized to ascertain the N-terminal sequence with Biotools software version 3.2, revealing 10 residues i.e., MTAVNQTISK having m/z ration of 1092.57 m/z (calculated) and 1092.381 m/z (measured). This is verified by identifying the peaks and examining the recombinant collagenase sequence coverage at the MS level, as depicted in Fig. 5A1, 5A2 and S1, respectively. Based on previous recommendations, it is emphasized that achieving a minimum of 4 amino acids and 20 % sequence coverage is essential for successful protein identification via PMF, that fortunately served the study purpose. However, this method did not uncover the entire N-terminus due to its reliance on trypsin, which cleaves the protein at specific R and K sites. Therefore, to authenticate the identification of the N-terminal sequence of the recombinant collagenase, a straightforward and functional cyclic method involving automated Edman degradation using a protein sequencer was utilized. This approach allows for the acquisition of longer (>15–30) amino acid residues at N-terminus. The sensitivity of Edman degradation enables the detection of peptides in quantities as low as 5 pmol. The present study established the N-terminal sequence, consisting of 30 amino acid residues, exhibiting a 99.76 % similarity to U32 family peptidase, as previously noted. This signifies that the initial 4 amino acids identified using Edman’s approach correspond with the MALDI-TOF-MS analysis, emphasizing the reliability and potential of this technique.
3.5.3 CD spectroscopy
CD spectroscopy helps understand protein and peptide structures, revealing α – helix, β – sheet and random coils in their secondary structures. The CD spectrum depicting the collagenase protein is illustrated in Fig. 6A. The spectrum displayed a negative ellipticity ranging from 220 − 230 nm, with a peak observed at 225 nm, signifying the presence of a proteins β structure. In a Tris HCl buffered solution, the CD spectrum of the collagenase protein exhibits distinct negative peaks, strongly suggesting the existence of β – sheet conformation. The analysis revealed a composition of 8.02 % α – helix and 22.14 % β – sheet content. Brito et al., 2021, reported collagenase activity from gold immobilized bromelain and the CD analysis data showed the presence of 24.2 % antiparallel β – sheet and 24 % α – helix showing bromelain belongs to α + β protein class. Indra et al., 2005, reported a broad α – helical secondary structure in molar ellipticity range of 208 – 222 nm for collagenase and in presence of Zn2+ ellipticity was reveled at 215 nm and further addition of Zn2+ caused a shift from α – helical to β – sheet structure after 215 nm, this shift in CD spectrum indicated a conformational change in the polypeptide backbone induced by Zn2+. Das et al. (2018), reported presence of both α – helix and β – sheet structures in broad negative troughs at 210 nm and 222 nm showing that the gelatinolytic protease was a type of α/β class of protein with well integrity in secondary and tertiary structure. CD spectrum of sedolisin, a subtilisin like protease, indicated the folded protein had a mixed α/β structure displaying broad negative spectra with two major peaks at ∼ 210 nm for α- helix and at ∼ 225 nm for β- sheet structure (Latka et al., 2015).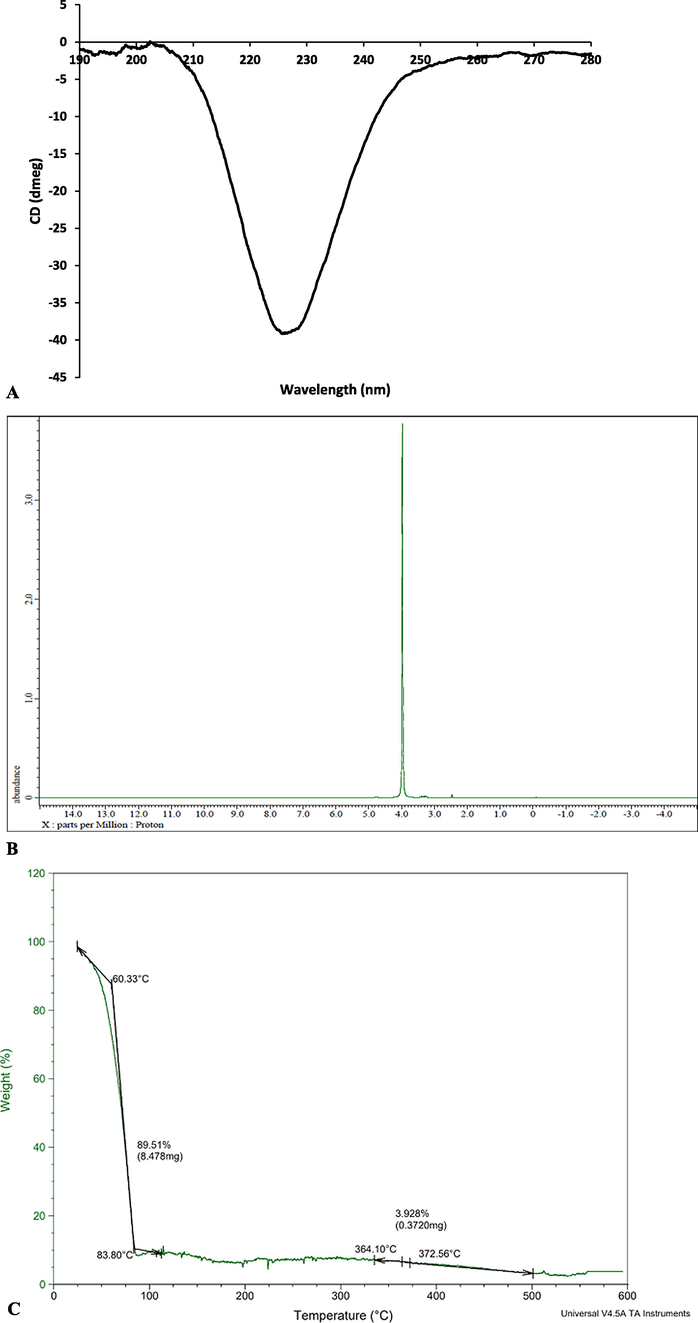
Analytical characterization of purified recombinant collagenase. (A)Circular dichroism spectra (B) 1H −NMR spectra (C) TGAanalysis.
3.5.4 1H NMR spectroscopy analysis
1H NMR was used to further characterize the structural characteristics of the recombinant collagenase (DMSO, 400 MHz, δ ppm) as illustrated in Fig. 6B. The characteristic signal of glycine and hydroxyproline are evidenced at δ = 4.0 ppm. A very narrow signal at δ = 2.4 ppm is visualized (Nimptsch et al., 2011). The peaks observed in 1H NMR spectra for collagenase was similar with spectra observed in literature. Do et al., 2023, observed proton NMR peak at δ = ∼ 4.0 ppm for collagenase treated microcapsules. Similar peaks in 1H NMR spectra of collagenase displayed by other proteases were reported in literature (Chitra Latka et al., 2015; Senyay-Oncel et al., 2014; Nithyapriya et al., 2021).
3.5.5 TGA analysis
TGA analysis of purified recombinant collagenase was verified for its thermostability performance. Two main stages of weight loss at particular temperatures were observed in TGA thermogram depicted in Fig. 6C. Initially maximum weight loss of 89.5 % occurred in temperature range between 25 °C to 125 °C due to loss of water molecules from protein and dehydration process. Additional weight loss of 3.928 % occurred between temperatures of 330 °C and 500 °C, attributed to the breakdown of amino acid residues, protein structure and bond cleavages. The thermal stability of recombinant collagenase was found to be upto 500 °C. The protein structure, its composition and conformation of the influencing factor for thermal stability. Massaro et al., 2023, reported similar thermal behavior of recombinant collagenase nanomaterials for degradation of organic matter present in nanomaterials. In consent to present research Călin et al., 2017, reported first zone (0––150 °C) is due to water evaporation and the second phase (150–––500 °C) is complete denaturation and organic degradation of polypeptide chains of keratin microfibrils and after 500 °C is complete degradation and obtaining carbonized residue. Mohapatra et al., 2020, revealed degradation of PHA produced from Bacillus megaterium QUAT016 in two stages with steep decrease in TGA curve as observed in present study.
3.6 Gelatin liquefaction test
The gelatin liquefaction test was conducted to evaluate the potential applicability of purified recombinant collagenase in gelatin degradation. The purified collagenase on 1 % agar test tube with 0.4 % gelatin showed partial liquefaction, forming a distinct liquefied zone and for 2 % agar test tube with 0.4 % gelatin, purified collagenase displayed partial liquefaction lesser than the either concentration (Fig. 7A, 7B). The observed liquefaction of gelatin by purified recombinant collagenase, in 1 % agar test tube, indicated significant liquefaction activity in degrading gelatin substrates. This degrading activity is crucial in various fields such as food processing, pharmaceuticals and biomedical research where gelatin is used as substrate. The differences in liquefaction among two concentrations of gelatin suggest variations in the purified collagenase effectiveness or specificity. This variability may have implications for the practical applicability of the isolates in specific gelatin degradation processes. In accordance to current study, researchers Batroukha et al., 2022 and Joseph et al., 2019, reported gelatin liquefaction ability of novel Lentzea sp. and Bacillus sp. as screening for identifying gelatinolytic activity. Yasmeen et al., 2022 carried out gelatin liquefaction from bacterial species isolated from fishes and have shown good gelatin degrading ability and that extracted gelatin can be further used in wide applications such as, in pharmaceutical products for soft and elastic gelatin capsules, useful in coatings like posters, glossy pages of magazines, food coatings as preservatives and also useful in cosmetics like lotions, creams and hair preparations.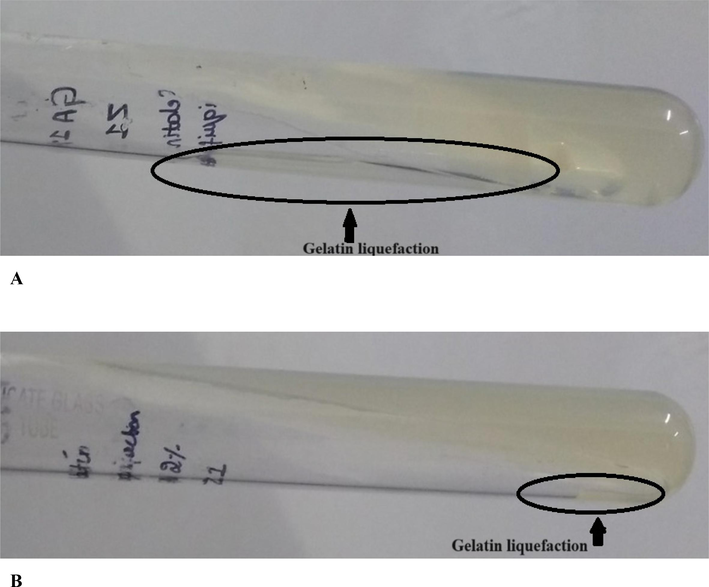
Gelatin liquefaction ability of Collagenase from Bacillus siamensis strain Z1. (A) 1% Agar- Gelatin test tube (B) 2% Agar − Gelatin test tube.
3.7 In-silico studies
3.7.1 Structural and functional characterization of collagenase
ExPASy translate tool was employed for translating nucleotide collagenase gene sequence to amino acid sequence. The Expasy translate tool provided six translated protein sequences of the submitted collagenase gene sequence. The result of Expasy translate provided (SI – S2A) clearly suggesting the first result provides a long-read continuous protein sequence of the subjected collagenase sequence whereas, the other sequences results are not useful due to their discontinuity and multiple ORFs. In line with this research, ExPASy translate tool was employed by other researchers for translating protease nucleotide sequences (Mushtaq et al., 2021; Perfumo et al., 2020).
The collagenase gene nucleotide phylogenetic classification was carried out using nucleotide sequences from different Bacillus species with accession numbers – Bacillus velezensis strain DH8043 chromosome complete genome (CP047268.1), Bacillus siamensis strain YB-1631 chromosome complete genome (CP110268.1), Bacillus amyloliquefaciens strain 205 chromosome complete genome (CP054415.1), Bacillus velezensis strain EN01 chromosome complete genome (CP053377.1) and Bacillus amyloliquefaciens strain R8-25 chromosome complete genome (CP054479.1). The result of the phylogenetic analysis showed that collagenase gene nucleotide sequence of Bacillus siamensis strain Z1 was closer to Bacillus amyloliquefaciens strain R8-25 chromosome complete genome, but slightly differed from Bacillus velezensis strain DH8043, Bacillus siamensis strain YB-1631, Bacillus amyloliquefaciens strain 205 and Bacillus velezensis strain EN01 as provided (SI-S2B). Phylogenetic tree analysis of collagenase gene protein sequence reveals that collagenase U32 family peptidase Bacillus siamensis is closely related to U32 family peptidase Bacillus siamensis (WP_064777716.1) followed by other U32 family peptidases namely Bacillus amyloliquefaciens (WP_045510354.1), Bacillus siamensis (WP_198697931.1), Bacillus sp. JFL 15 (WP_049627089) and far related to U32 family peptidase Bacillus nakamurai (WP_061522884.1) provided (SI-S2C). The neighbor joining algorithm was applied in generating phylogenetic trees (SI- S2B, S2C) that employs the computed distance matrix to iteratively merge sequences into tree structure by minimizing the total branch lengths. Phylogenetic analysis of collagenase protein sequence was also reported by Rani and Pooja (Rani and Pooja, 2018), A phylogenetic tree was built using nine collagenase protein sequences obtained from various Pseudomonas species, showcasing their close evolutionary relationship. The study also highlights the limited applicability of collagenase from pathogenic bacterial species, such as Clostridium histolyticum, in the food industry due to safety concerns. This underscores the potential role of collagenase from non-pathogenic bacterial species to address this limitation. Phylogenetic tree of protein sequence of MMP from Lucilia sericata larvae showed a notable connection among insect MMPs. Additionally, human and insect MMPs differed significantly from bacterial MMPs. Human MMP-1 shared a close relation with insect MMP-1(Alipour et al., 2017).
The collagenase gene protein sequence was subjected to pair wise alignment with top similar sequence, wherein collagenase gene showed 100 % similarity with Bacillus sp. this sheds light on validation of collagenase gene belongs to U32 family peptidase Bacillus sp. and is illustrated in (SI-S2D). The current research identified the collagenase gene from Bacillus siamensis strain Z1 belongs to the Peptidase U32 family domain (SI-S2E). This determination was made using a scientific analysis online tool known as InterProScan. The tool revealed amino acids of range 93–––325 and 337–––419 of collagenase gene contributed for family and Domain characteristic of peptidase U32 respectively. The identified peptidase U32 family domain is recognized for its involvement in proteolytic activities and shares homology with similar sequences among other microbial collagenases. This finding shed light on the evolutionary lineage and functional characteristics associated with the collagenase gene of interest, offering insights into its potential biological roles and relationships within the broader peptidase domain family. In alignment with this study, Bhattacharya et al., 2019, utilized the InterProScan tool, revealing that the collagenase synthesized by Pseudomonas agarivorans NW4327 was categorized within the peptidase U32 domain. Novel keratinases from Bacillus cereus group. A keratinase gene, KerS revealed close resemblance to peptidases S8 thermitase like domain at 110–368 amino acids using InterProScan and NCBI conserved domains search (Almahasheer et al., 2022).
The physico-chemical features of collagenase gene from Bacillus siamensis strain Z1 was computed using ProtParam tool. The analysis of collagenase gene sequence revealed an amino acid composition by a notable prevalence of Glutamine (9.2 %), Alanine (8.5 %), Lysine (7.3 %) and Isoleucine (7.3 %) among other residues. The calculated molecular weight of the gene was determined to be 47,447.96 Da which is similar with the ∼ 47 kDa obtained by SDS-PAGE mentioned in section 2.3. The molecular formula is predicted to be C2112H3290N556O644S21 with 6623 total number of atoms. The instability index was computed to be 37.22, suggesting good stability, as protein indices below 40 is considered stable. Moreover, the Aliphatic index was found to be 82.06, that hints at potential thermostability, where higher values correspond to increased thermostability. The GRAVY score was calculated to be −0.295 suggesting the protein sequence is moderately hydrophilic. A negative GRAVY value signifies a prevalence of hydrophilic amino acids within the sequence. The extinction co-efficient, measured at 48860 M−1 cm−1 at 280 nm, signifies light absorption characteristics. The estimated half-life was calculated as 30 h in mammalian reticulocytes, in-vitro, >20 h in yeast, in-vivo and > 10 h in E. coli, in-vivo indicating favorable stability within a cellular environment. Bacillus cereus group keratinases were also assessed for physiochemical analysis by ProtParam tool. The analysis revealed gravy index ranging between −0.32 to −0.35 and 73.98 to 75.69 aliphatic index and 397 amino acids of protein sequences with molecular weight ranging between 42.2 – 42.4 kDa(Almahasheer et al., 2022). A serine protease AKD9 was analyzed for physiochemical properties by using ProtParam tool, the results revealed AKD9 comprising 442 amino acids possessed gravy index of −0.37, instability index of 30.63 which is less than 40 suggests that protein is stable and Aliphatic index of 79.84 (Mahmoud et al., 2021).
The SWISS-MODEL online server tool was employed to generate a structural model for the collagenase gene based on homology modelling. The model was constructed using the structure of tRNA hydroxylation protein P1 (O32034.1.A) as template due to its high sequence similarity of 93.13 % with the target protein. The 3D structure of the resulting model is illustrated in SI-S2F. Structural evaluations conducted via Ramachandran plot analysis revealed that 96.19 % of the residues were under favored region, 0.48 % as Ramachandran outliers (SI-S2G) and MolProbity score of 0.82 indicating good structure quality. The model’s overall quality was assessed through GMQE (Global Model Quality Estimation) score which yielded 0.94 indicating high confidence level in quality of the predicted collagenase gene model. Scores closer to 1.0 generally suggest better model reliability, with 1.0 being the highest score, indicating a very reliable prediction. Therefore, a GMQE score of 0.94 signifies a high-quality prediction in collagenase structure accuracy and reliability. The modelled collagenase gene structure had an overall quality factor of 95.41 by ERRAT analysis suggesting a good stable structure (SI – S2H). The secondary structure of collagenase gene can be noticed in Fig. 5B that illustrates MSA with related Bacillus species and also the presence of β-strands, α-helix, turns and coils. It observed that maximum of β – structures are present in modelled collagenase structure. Homology modelling for collagenase of Bacillus cereus using SWISS MODEL was reported by Abfalter et al., 2016, stating with sequence identity of 49.80 % observed between B. cereus ATCC 14579 ColA and the chosen template. The quality of model was ascertained by RAMPAGE and 95.8 % residues in favored region, 3.1 % in allowed region and 1 % as outliers. Homology modelled structure of serine protease SAPRH was built using subtilisin NAT as template with 71.6 % similarity between sequences and the model Ramachandran showed most of the residues in favored region (Rekik et al., 2019). Similarly, a serine protease SAPB 3D structure was built Subtilisin E as template which had 69 % sequence identity, and it was reported Leu31 and Thr33 were the important residues of active site participating in substrate recognition for catalysis reaction and animal hide depilation (Zaraî Jaouadi et al., 2014). The predicted secondary structure of AKD9 serine protease indicated a composition comprising α-helix, β-strands, β-turns and dominance of coils in structure (Mahmoud et al., 2021).
The inherent variability of proteins, particularly in their high-flexible loop structures, is crucial for enzyme catalysis. However, these loops are often susceptible to heat and tend to be the initial structures to unfold under extreme conditions. In this research, FoldUnfold was used to forecast the regions of high flexibility and disorder within the collagenase gene. The tool identified 1 unfolded region consisting of 11 amino acids (164–174 aa) and depicted in SI-S2I. Su et al., 2022, reported the usage of FoldUnfold to predict highly flexible disordered regions of keratinase KerBp.
3.7.2 Molecular docking study
The in-silico molecular docking method was employed to identify the affinity between the collagenase protein and chosen substrates (ligands) such as APHCP, Collagen type 4 alpha (531–543), PZ-peptide and FALGPA. The 2D, 3D structures of substrates utilized for docking along with their PubChem ID is illustrated in SI-S3. This study employed two distinct tools namely, PyRxautodock vina and classic Autodock, for performing molecular docking analysis. In PyRx for blind molecular docking grid box was designed using co-ordinates center X=3.8971, Y=6.7699 and Z=-4.3388 that covered the whole collagenase protein. By employing PyRx software docking score of −8.1 kcal/mol was observed for substrate APHCP by 7 hydrogen bonds with amino acids such as PHE 341, ASN 361, LYS 270, MET 269 and ASN 311 (Fig. 8A). Followed by −7 kcal/mol for PZ-peptide substrate with ARG 360, PHE 341, ASP 340, PHE 339, THR 337 and SER 271 by 6 hydrogen bonds (Fig. 8C). A docking score of −6.8 kcal/mol was obtained for collagen type 4 alpha substrate with 13 hydrogen bonds with interaction for residues LYS 79, TYR 81, ARG 155, GLU 177, GLU 23, ASP 287, ARG 283, PRO 374, ILE 376, ASN 378 and GLU 305 (Fig. 8B). A docking score of −6.7 kcal/mol was derived for FALGPA substrate with interaction with amino acids such as ASN 311, THR 337, PHE 341, ASN 361 and ASP 340 with 5 hydrogen bonds (Fig. 8D). The Hydrogen bond interactions along with interaction distance analyzed for docking by PyRx software is provided in SI-S4.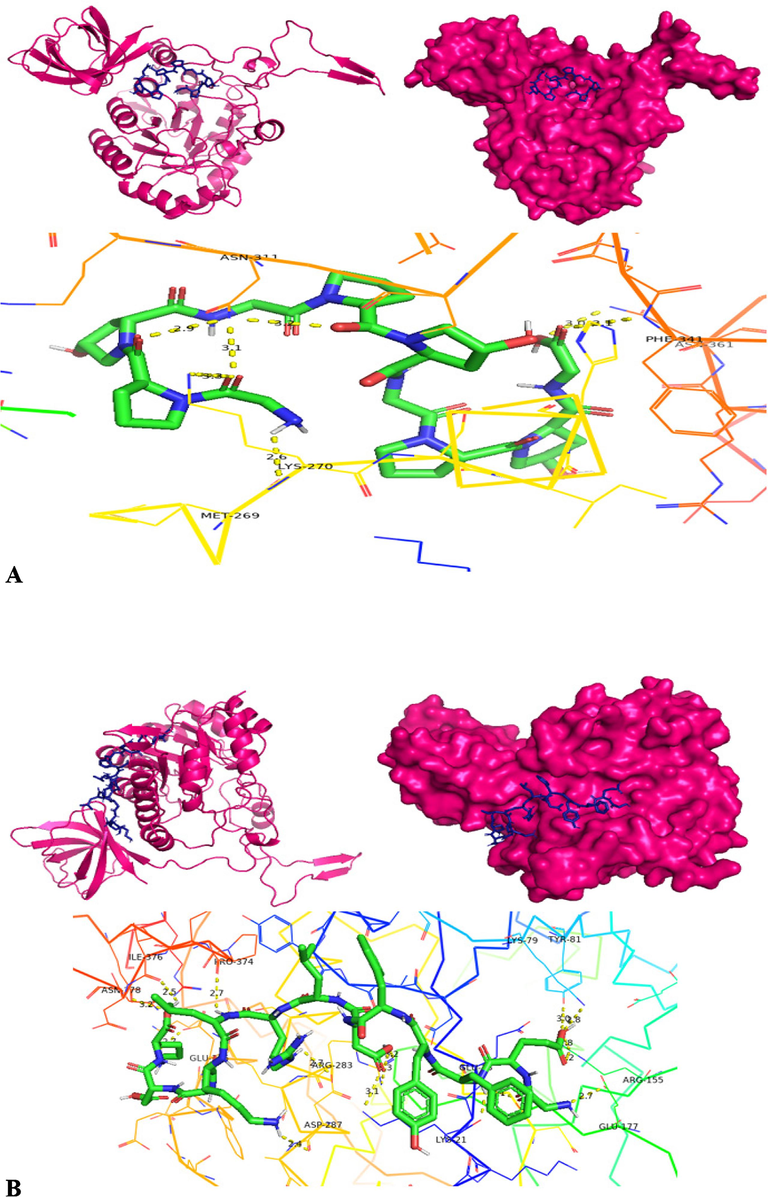
Molecular docking structures – cartoon diagram, surface diagram, H-bond interactionsbyPyRx software docking of collagenase with different substrates- A) APHACP (Alaska pollock hydroxyproline containing marine collagen peptide), B) Collagen type 4 alpha (531–543), C) PZ – peptide and D) FALGPA (Pink – Protein; Blue – Substrate in cartoon and surface diagram; Hydrogen bond interaction diagram: Green – Substrate, Yellow dots – Hydrogen bond, Colored Ribbon structure – Collagenase protein).
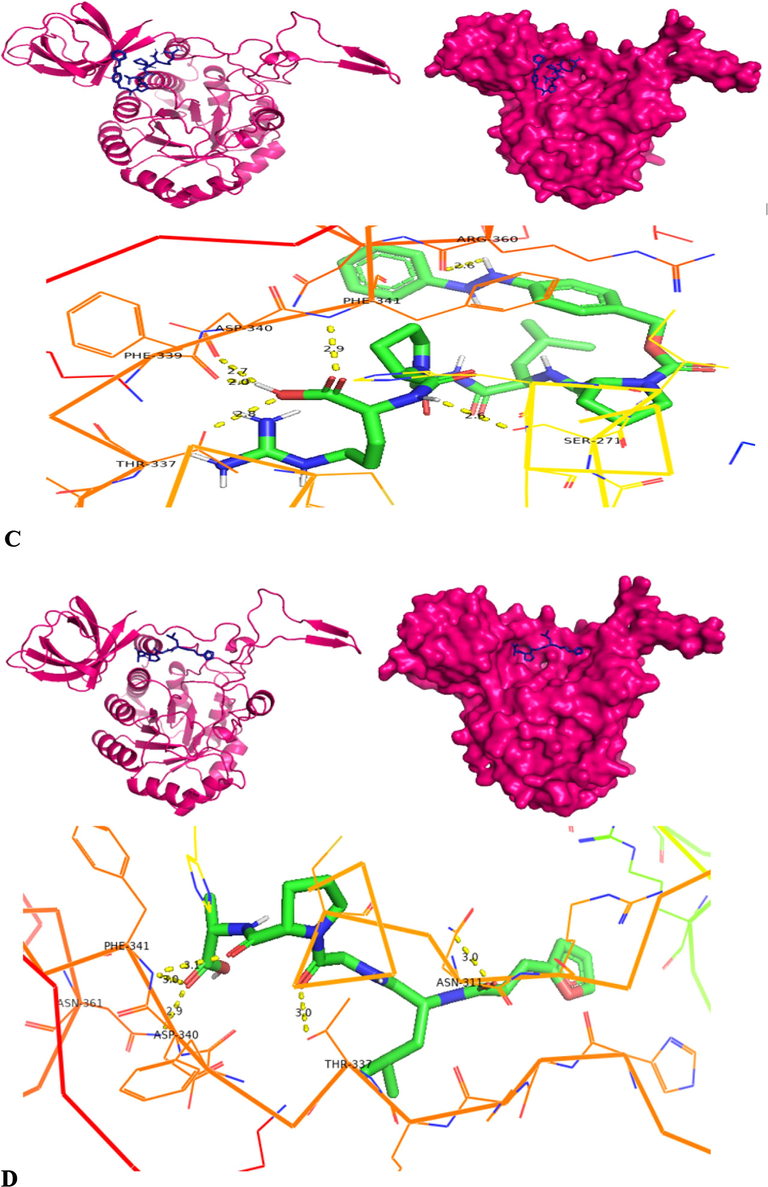
Molecular docking structures – cartoon diagram, surface diagram, H-bond interactionsbyPyRx software docking of collagenase with different substrates- A) APHACP (Alaska pollock hydroxyproline containing marine collagen peptide), B) Collagen type 4 alpha (531–543), C) PZ – peptide and D) FALGPA (Pink – Protein; Blue – Substrate in cartoon and surface diagram; Hydrogen bond interaction diagram: Green – Substrate, Yellow dots – Hydrogen bond, Colored Ribbon structure – Collagenase protein).
Molecular docking by autodock software included the protein preparation for ligand–protein docking by removing excess water molecules, integrating polar hydrogen atoms, merging non-polar hydrogen atoms, and addition and dispersion of charge. The processed protein and ligand molecules are saved in.pdbqt format using Autodock software. The grid co-ordinates used in PyRx were employed in autodocking to known and compare the best docking score obtained from two different docking tools. The docking process is conducted for top 10 best poses of collagenase protein – ligand complex. The configurations are chosen based on the ligand binding energy cluster results and number of hydrogen bonds.
According to the docking results, collagenase protein interacts with active amino acids such as ARG 360, CYS 309, ASN 311, ARG 312, HIS 333, ARG 215, GLN 212 by making 10 hydrogen bonds with substrate APHCP(Fig. 9A) that contains a marine collagen peptide and resulting in highest docking score of −12.7 kcal/mol. Followed by next docking score of −9.6 kcal/mol with FALGPA substrate by 4 hydrogen bonds with PHE 341, SER 271 and ASN 311 (Fig. 9D). The binding complex of collagenase with PZ-peptide resulted in binding score of −8.7 kcal/mol forming 6 hydrogen bonds with SER 271, HIS 273, ASN 311 and HIS 333 (Fig. 9C). Collagenase was bonded with 8 hydrogen bonds with amino acid residues such as GLU 266, SER 271, HIS 273, ARG 55, THR 337, LYS 335 and ASN 311 of collagen type 4 substrate resulting in lowest docking score of −7.7 kcal/mol (Fig. 9B) by utilizing autodock software. The Hydrogen bond interactions along with interaction distance analyzed for docking by autodock software is also provided in SI-S4.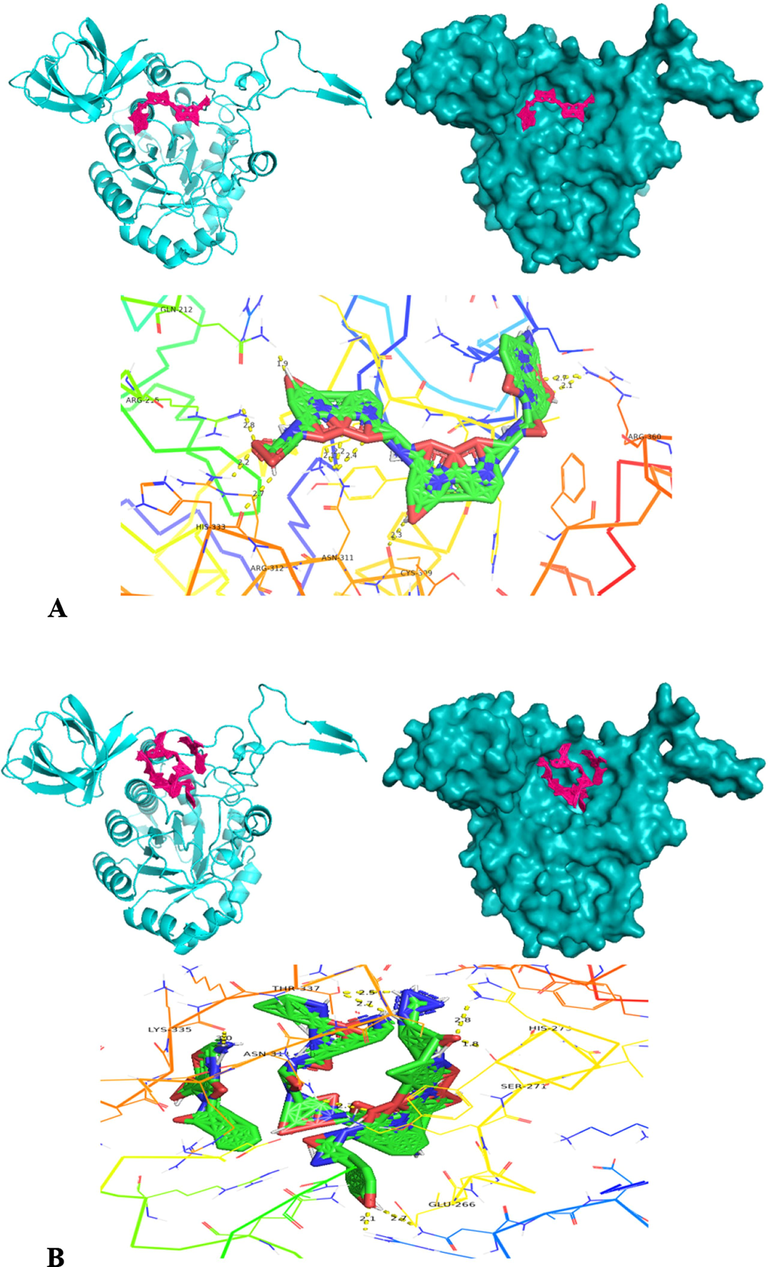
Molecular docking structures – cartoon diagram, surface diagram, H-bond interactionsbyAutodock software of collagenase with different substrates- A) APHACP (Alaska pollock hydroxyproline containing marine collagen peptide), B) Collagen type 4 alpha (531–543), C) PZ – peptide and D) FALGPA (Cyan – Protein; Pink – Substrate in cartoon and surface diagram; Hydrogen bond interaction diagram: Green – Substrate, Yellow dots – Hydrogen bond, Colored Ribbon structure – Collagenase protein).
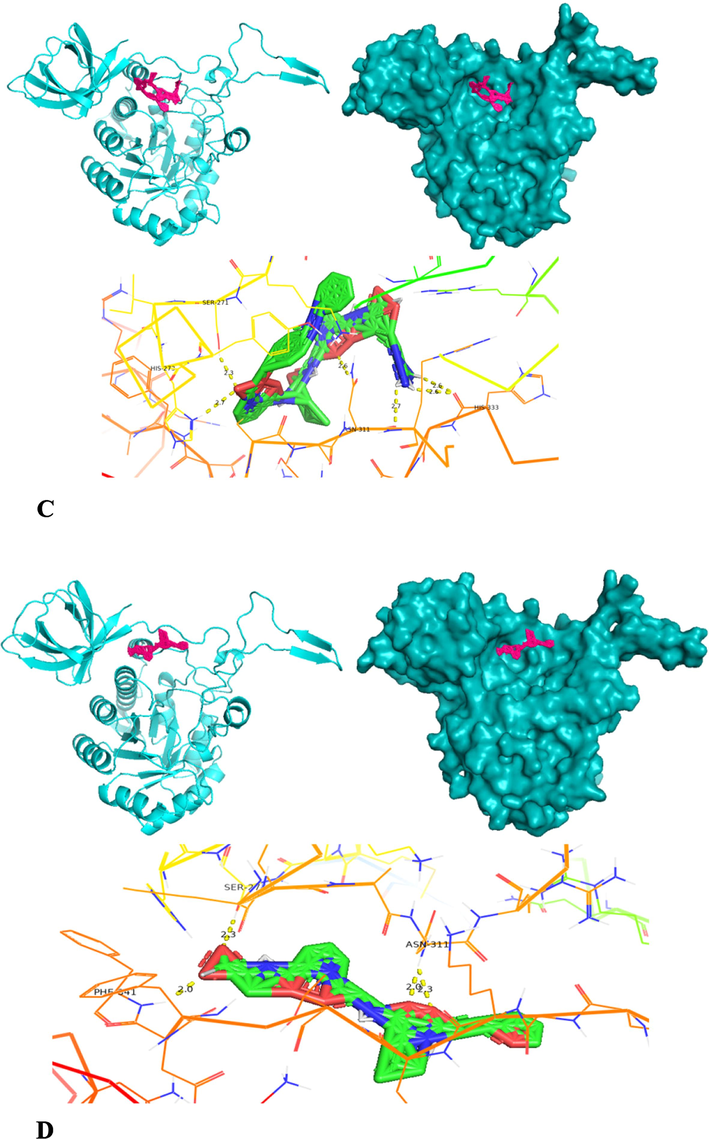
Molecular docking structures – cartoon diagram, surface diagram, H-bond interactionsbyAutodock software of collagenase with different substrates- A) APHACP (Alaska pollock hydroxyproline containing marine collagen peptide), B) Collagen type 4 alpha (531–543), C) PZ – peptide and D) FALGPA (Cyan – Protein; Pink – Substrate in cartoon and surface diagram; Hydrogen bond interaction diagram: Green – Substrate, Yellow dots – Hydrogen bond, Colored Ribbon structure – Collagenase protein).
The insights gained from molecular docking results elucidate the involvement of numerous amino acids in the interaction of collagenase with diverse substrates. Among them, amino acids including SER 271, HIS 333, ASN 311, HIS 273, THR 337, PHE 341, ARG 360 and ASN 361 consistently reoccur in the docking outcomes across four substrates. Hence it is assumed that these specific amino acids play a contributory role in establishing the active site within the collagenase protein. This active site facilitates the interaction and binding of various substrates to the protein and aids in the catalytic reaction.
In the present research study, the collagenase activity was determined using collagen substrate derived from extrapure marine fish, 95 % purity. Through molecular docking analysis, it became evident that collagenase docking with APHCP, containing marine collagen peptide, yielded the highest docking score of −12.7 kcal/mol. This observation serves as validation for both wet lab experiments and in-silico studies. Furthermore, the collagenase-APHCP docked complex is subjected to molecular dynamics and simulations. These simulations aim to comprehensively investigate the dynamics, interactions, and alterations occurring between the protein and substrate under various conditions and within different environment. Mechri et al., 2022, reported blind molecular docking of synthetic substrate N-succinyl-L-Phe-L-Ala-L-Ala-L-Phe-p-nitroanilide into active site of modelled serine protease from Streptomyces mutabilis strain TN-X30 and it revealed the 21 amino acids involvement in substrate binding with binding energy −4.3174 kcal/mol and serine residue of catalytic triad was considered as main residue in substrate catalysis. A similar study of substrate docking using N-succinyl-L-alanyl-L-alanyl-L-prolyl-L phenylalanine 4 nitroanilide S-9205 against mutant and wild keratinases. Mutant proteins D137N of KerS13 and 7 mutants of KerS26uv keratinases and wild protein exhibited affinity score of −7.17, −7.43 and −6.57 (Almahasheer et al., 2022). Enzyme −substrate docking was also experimented with serine protease and ligands by Rekik et al., 2019, wherein homology modelled serine protease SAPRH from Bacillus safensis RH12 was docked with suc-AAPF-pNA, suc-AAPV-pNA and suc-ALPA-pNA resulting with interaction energy of −134, −106 and −123 kJ/mol, respectively using MOE docking software.
3.7.3 Molecular dynamics and simulations analysis
Molecular dynamics study was conducted on apo protein (modelled collagenase gene) (APO) and collagenase protein docked with substrate (APHCP) considered as ligand (DRG1) for 200 ns simulation run using GROMACS system. The method of molecular dynamics simulation aids in force calculations and understanding amino acid atom motion. Considering the various factors like RMSF, SASA, Rg and RMSD and interactions with hydrogen bond the stability of the docked complex is evaluated. The RMSD factor helps in understanding the stability of ligand when complexed with target protein. PRODRUG server (https://davapc1.bioch.dundee.ac.uk/cgi-bin/prodrg/submit.html) is employed to generate the ligand-topology file that contains ligand in complex with the protein. The protein’s PDB file, featuring the APO state and the protein complexed with ligand (APHCP), is uploaded to the PRODRG server. The resultant energy-minimized topology file obtained from this process is utilized for simulations via gromacs.
The RMSD plot is created to ascertain the stability of the docked and APO collagenase protein. Fig. 10A depicts the RMSD plot for APO and protein-substrate complex (DRG1). In the plot it is observed that for initial 60 ns fluctuations have occurred for APO protein and after 60 ns it is constant and stable, whereas for ligand fluctuations are observed throughout 200 ns run. The average RMSD value is predicted to be ∼ 0.523 ± 0.32 nm for APO protein and ∼ 0.615 ± 0.48 nm for collagenase protein and APHCP. The RMSD plot indicated the relative stability of the APO and Protein-Ligand complex in a system from 70 ns to 150 ns. RMSF evaluates amino acid residue consistency and divergence throughout the simulation. Fig. 10B illustrates the RMSF plot depicting amino acid residues of collagenase protein APO and protein-ligands [Collagenase and APHCP] complex. A lower RMSF (Root Mean Square Fluctuation) value indicates a stable enzyme – substrate complex, whereas a higher RMSF value suggested increased flexibility in dynamic simulations. In the RMSF plot it can be seen that both APO and docked complex have shown fluctuations for amino acids ranging from 180 to 240 and also the docked complex showed higher RMSF value upto 1.6 nm and 0.8 nm for APO protein. But in overall RMSF plot, lower RMSF in range of 0.2 to 0.4 nm is observed in ligand (APHCP substrate) molecule within the collagenase protein complex indicating its increased stability, demonstrating its equilibrium with COX-2 over the 200 ns simulation. According to the current study outcome, when an enzymes flexibility decreases it undergoes conformational changes that facilitate substrate binding during catalysis. Using the RMSF of the enzyme – substrate complex, the catalytic potential of modelled collagenase and APHCP was investigated.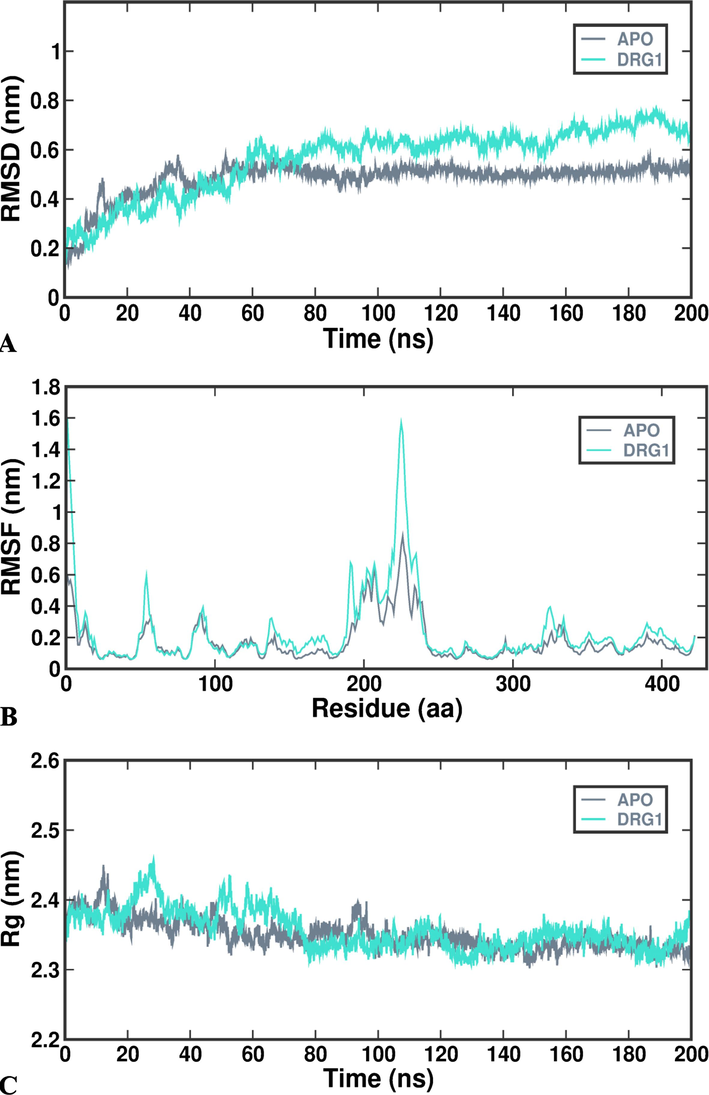
Molecular dynamics and simulations of docked complex- (A) RMSD; (B) RMSF; (C) Rg; (D) SASA; (E) Intra-Hydrogen bond.
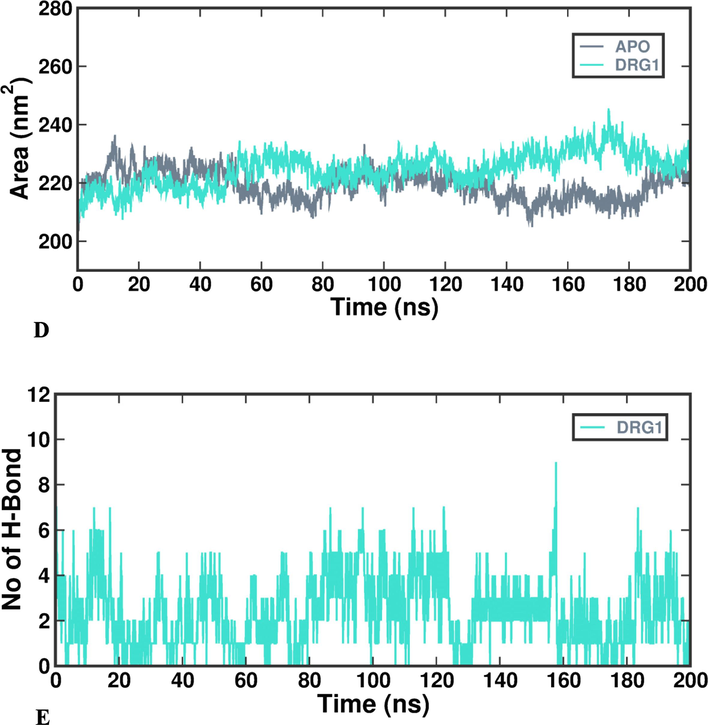
Molecular dynamics and simulations of docked complex- (A) RMSD; (B) RMSF; (C) Rg; (D) SASA; (E) Intra-Hydrogen bond.
Rg values depict protein rigidity and compactness in the molecular dynamics system during ligand interaction. APO and collagenase protein in complex with APHCP are found to have typical Rg values between ∼ 2.378 ± 0.53 nm and ∼ 2.397 ± 0.21 nm for 200 ns of simulation time as depicted in Fig. 10 C. Enzyme folding indicates the stability of the Rg, while Rg fluctuations indicate the unfolding of the enzymes. Throughout the simulation, substrate APHCP consistently exhibited a stable and unchanging Rg value. This constancy points to the substrate’s enduring compactness and structural stability. Such unvarying alignment suggests the substrate’s precisely maintained conformation, providing evidence of its resilience and uniform performance over the whole 200 ns trajectory. SASA is measured to determine the protein modulation within the system. SASA of the complex remained constant and equilibrated during the simulation, and stability was consistent over 200 ns of simulation. The SASA for Collagenase protein (APO) and docked complex (DRG-1) is obtained as ∼ 220 ± 7.56 nm2 and ∼ 225 ± 6.89 nm2. Assessing the SASA alteration measures the extent of complex protein aggregation in the system. Fig. 10D displays the SASA plot for Collagenase – APO and APHCP. The analysis of hydrogen bonding within the modelled collagenase gene and APHCP complex reveals a continuous binding interaction of APHCP with collagenase protein throughout the duration. More than 8 pairs of hydrogens are stable for the docked complex. Throughout the 200 ns simulation, the continuous presence of hydrogen bonds substantiates the stability and equilibrium of APHCP in complex with collagenase protein. The depiction of hydrogen bonds formed during the MD simulations is illustrated in Fig. 10E. The analysis of RMSD, RMSF, Rg and SASA plots collectively demonstrates the good binding of APHCP within collagenase protein binding site, maintaining stability throughout the entire 200 ns simulation duration. Similar to current study, Bhattacharya et al., 2019, reported the MDS of modelled U32 collagenase from Pseudomonas agarivorans NW4327. In the study MDS was carried out using GROMACS system for modeled apocollagenase and collagenase structures with triple helical collagen peptide fragment and complexed with calcium bound and unbound. A smaller deviation was noticed between RMSD values of apocollagenase and collagenase complex with collagen and metal ion and it was stated smaller the deviation, the more spatially equivalent the two states (starting and stimulated) of the proteins were and the more stable the protein structure.
In similar context of current research, Bhatt et al., 2021, showed molecular docking of enzyme and substrate with respect to glyphosate oxidoreductase and C-P lyase that are main enzymes in bioremediation of glyphosate. Redocking was performed to estimate accuracy of PyRx. The docking analysis of Glyphosate oxidoreductase (GOX)with glyphosate reveals a binding score of −5.4 kcal/mol and that with C-P-lyase was −4.3 kcal/mol and the complex of GOX-glyphosate was chosen potential for dynamics study. GOX-glyphosate was most stable during MD simulation, with an RMSD of 0.62 nm and little fluctuations noticed in RMSF, and Rg plot showed that enzyme substrate complex exhibited stable compactness and six hydrogen bonds were observed between GOX – glyphosate complex. Similarly, to current research Rahimnahal et al., 2023, generated a 3D model for keratinase and verified with Ramachandran plot, ERRAT score, Verify-3D showing 90 % amino acids in favored region, protein–protein molecular docking was performed with chicken feather keratin 4, 12. Further Molecular dynamics (MD) simulations showed keratinase KRLr1 interacted strongly with chicken FK-4 than chicken FK-12 for 50 ns duration with serine 175 as key amino acid in interaction and maintaining more than eight hydrogen bonds with modelled keratinase.
3.7.4 MMPBSA analysis
The binding efficiencies of collagenase with APHCP was identified by using MM-PBSA calculation. The evaluation of disparities and post-simulation rationalization of docked complexes is executed through MMPBSA analysis. The assessment of ligand affinity is determined within the protein binding core where interactions occur. The stability of the protein–ligand complex heavily relies on binding energy, where negative energy signifies increased stability (Revankar et al., 2023). Fig. 11. Illustrates data, graph from MMPBSA calculations centered on active site binding interactions. The results suggest that GLN212, ARG215, ASN311, ARG312, HIS333 and ARG360 amino acid residues in collagenase gene play a significant role, contributing most to total binding energy of −7.9269 kJ/mol. Active site residues contributed to H-bond formation and were evaluated by MMPBSA pocket analysis. Molecules maintained consistent positions throughout the simulation.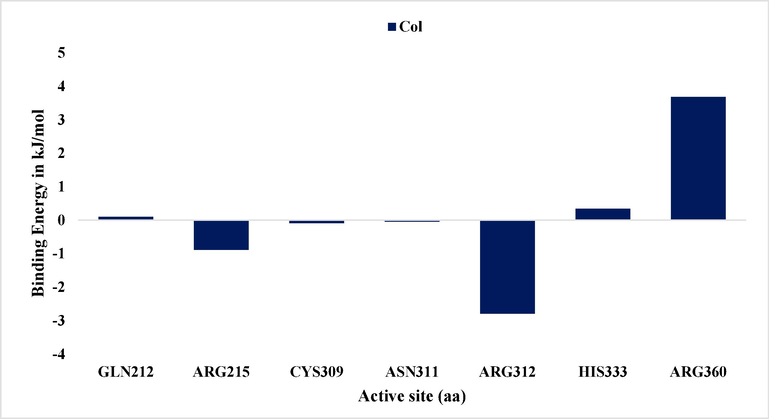
The Interaction energy at the active site (Binding energy) determined by MM-PBSA in the simulation of the highest docked score complex.
Singh and Kumar, 2023, reported a good ligand–protein binding energy in MMGBSA analysis and the Molecular dynamics simulation trajectory analysis showed significant stable and energetically favorable binding of Naringin dihydrochalcone (NDC) at the catalytic site of MMP-2. MMPBSA binding energy for GOX and glyphosate complex was found in acceptable range and the results showed GOX-Glyphosate complex is stable and GOX was found to be effective for bioremediation of glyphosate. The per residue interaction involved in GOX-Glyphosate complex for MMPBSA analysis at 20 ns had negative and positive residues and higher negative per residue interactions showed these amino acids had higher affinity for glyphosate in catalytic reactions (Bhatt et al., 2021).
4 Conclusion
Collagenase gene was identified from Bacillus siamensis strain Z1 and development of recombination by cloning, expression, affinity purification. The molecular mass of purified collagenase was ∼ 89.4 kDa. The recombinant collagenase exhibited stability with numerous biochemical factors such as optimal pH and temperature, metal ions, thermodynamic properties, catalytic efficiency etc. and possesses alkaline thermostability. Collagenase identity was validated using MALDI-TOF-MS, while the structural elucidation by CD, 1H NMR and thermostability by TGA. The structural and functional characterizations such as homology modelling, family domain identification, physiochemical properties and phylogenetic analysis of collagenase gene was carried out using numerous computational tools. Molecular docking of collagenase with four different substrates using two docking software (PyRx and Autodock) was employed. The highest docked scored −12.7 kcal/mol of complex comprising modelled collagenase and substrate APHCP was analyzed for molecular dynamics and simulations for 200 ns.
Author contributions
Theconceptualization of research was carried out by AR and ZB. AR, GD and NB have undertaken experiments on gene cloning, expression and purification. AR and ZB were involved in biochemical and molecular characterization and in-silico studies. AR and GD have carried out MALDI analysis. AR and ZB accomplished data evaluation and interpretations and has written the original manuscript draft. ZB, IS, NM, AK and BM have reviewed the manuscript and provided critical inputs. All authors approved the manuscript's contents. ZB has supervised the entire research work.
CRediT authorship contribution statement
Archana G. Revankar: Conceptualization, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. Zabin K. Bagewadi: Conceptualization, Data curation, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. Ibrahim Ahmed Shaikh: Formal analysis, Funding acquisition, Writing – review & editing. G Dhananjaya: Methodology, Validation. Nilkamal Mahanta: Data curation, Formal analysis, Validation. Aejaz Abdullatif Khan: Formal analysis. Neha P. Bochageri: Investigation, Methodology, Validation. Basheerahmed Abdulaziz Mannasaheb: Data curation, Formal analysis.
Acknowledgements
The authors are pleased to express their gratitude to KLE Technological University, Hubballi, for providing support under the Ph. D Fellowship Program. The authors would like to acknowledge Department of Biological Sciences, Indian Institute of Science, Bangalore for MALDI-TOF-MS and application team, mass spectroscopy division, Bruker India Scientific Pvt. Ltd for N-terminal data acquisition. The authors are thankful to the Deanship of Scientific Research at Najran University for funding this work under the Research Group Funding program grant code NU/RG/MRC/12/14.The authors would like to express sincere gratitute to AlMaarefa University, Riyadh, Saudi Arabia for supporting this research.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Protease immobilization on a novel activated carrier alginate/dextrose beads: Improved stability and catalytic activity via covalent binding. Int. J. Biol. Macromol.. 2023;230:123139
- [CrossRef] [Google Scholar]
- Cloning, Purification and Characterization of the Collagenase ColA Expressed by Bacillus cereus ATCC 14579. PLoS One. 2016;11:e0162433.
- [Google Scholar]
- Purification and characterization of a new thermophilic collagenase from Nocardiopsis dassonvillei NRC2aza and its application in wound healing. Int. J. Biol. Macromol.. 2018;116:801-810.
- [CrossRef] [Google Scholar]
- Quantum modelling and molecular docking evaluation of some selected quinoline derivatives as anti-tubercular agents. Heliyon. 2020;6:e03639.
- [Google Scholar]
- Microbial bioremediation of feather wastes and ecofriendly production of keratinase and collagenase enzymes by Didymella keratinophila, the firstrecorded fungus from Assiut. Egypt 2022
- [CrossRef] [Google Scholar]
- Identification, Isolation, Cloning, and Expression of a New Alkaline Serine Protease Gene from Native Iranian Bacillus sp. RAM 53 for Use in the Industry. Appl. Biochem. Biotechnol.. 2023;195:5730-5746.
- [CrossRef] [Google Scholar]
- Therapeutic applications of collagenase (metalloproteases): A review. Asian Pac. J. Trop. Biomed.. 2016;6:975-981.
- [CrossRef] [Google Scholar]
- Molecular characterization of matrix metalloproteinase-1 (MMP-1) in Lucilia sericata larvae for potential therapeutic applications. Electron. J. Biotechnol.. 2017;29:47-56.
- [CrossRef] [Google Scholar]
- Expression of a New Recombinant Collagenase Protein of Lucilia Sericata in SF9 Insect Cell as a Potential Method for Wound Healing. Iran. J. Biotechnol.. 2019;17:e2429.
- [Google Scholar]
- Novel feather degrading keratinases from Bacillus cereus group: biochemical. Genetic Bioinform. Anal. Microorganisms. 2022;10:93.
- [CrossRef] [Google Scholar]
- Biochemical and molecular characterization of an acido-thermostable endo-chitinase from Bacillus altitudinis KA15 for industrial degradation of chitinous waste. Carbohydr. Res.. 2020;495:108089
- [CrossRef] [Google Scholar]
- One-pot synthesis and biochemical characterization of a magnetic collagenase nanoflower and evaluation of its biotechnological applications. Colloids Surf., B Biointerfaces. 2022;211:112302
- [CrossRef] [Google Scholar]
- Statistical optimization and characterization of bacterial cellulose produced by isolated thermophilic Bacillus licheniformis strain ZBT2. Carbohydr. Res.. 2020;491:107979
- [CrossRef] [Google Scholar]
- Bagewadi, Z.K., Mulla, S.I., Shouche, Y., Ninnekar, H.Z., 2016. Xylanase production from Penicillium citrinum isolate HZN13 using response surface methodology and characterization of immobilized xylanase on glutaraldehyde-activated calcium-alginate beads. 3 Biotech 6, 164. DOI: 10.1007/s13205-016-0484-9.
- Biochemical and enzyme inhibitory attributes of methanolic leaf extract of Datura inoxia Mill. Environ. Sustain.. 2019;2:75-87.
- [CrossRef] [Google Scholar]
- Production and Optimization of Gelatinase Producing Lentzea sp. Strain Isolated from the Soil Rhizosphere. Sci. J. Damietta Fac. Sci.. 2022;12:124-131.
- [CrossRef] [Google Scholar]
- Collagen and collagenolytic proteases: a review. Biocatal. Agric. Biotechnol.. 2018;15:43-55.
- [CrossRef] [Google Scholar]
- Binding interaction of glyphosate with glyphosate oxidoreductase and C-P lyase: Molecular docking and molecular dynamics simulation studies. J. Hazard. Mater.. 2021;409:124927
- [CrossRef] [Google Scholar]
- Cloning, Expression, and structural elucidation of a biotechnologically potential alkaline serine protease from a newly isolated Haloalkaliphilic Bacillus lehensis JO-26. Front. Microbiol.. 2020;11
- [CrossRef] [Google Scholar]
- U32 collagenase from Pseudoalteromonas agarivorans NW4327: activity, structure, substrate interactions and molecular dynamics simulations. Int. J. Biol. Macromol.. 2019;124:635-650.
- [CrossRef] [Google Scholar]
- Collagenase activity of bromelain immobilized at gold nanoparticle interfaces for therapeutic applications. Pharmaceutics. 2021;13:1143.
- [CrossRef] [Google Scholar]
- Degradation of keratin substrates by keratinolytic fungi. Electron. J. Biotechnol.. 2017;28:101-112.
- [CrossRef] [Google Scholar]
- Two steps purification, biochemical characterization, thermodynamics and structure elucidation of thermostable alkaline serine protease from Nocardiopsis alba strain OM-5. Int. J. Biol. Macromol.. 2021;169:39-50.
- [CrossRef] [Google Scholar]
- Purification, peptide mapping and spectroscopic characterization of myoglobin from striped snake-head fish (Ophicephalusstriatus) Orient. J. Chem.. 2016;32:181-194.
- [CrossRef] [Google Scholar]
- The gene expression omnibus database. Statistical Genomics. 2016:93-110.
- [CrossRef] [Google Scholar]
- Costa, E.P., Brandão-Costa, R.M.P., Albuquerque, W.W.C., Nascimento, T.P., Sales Conniff, A.E., Cardoso, K.B.B., Neves, A.G.D., Batista, J.M. da S., Porto, A.L.F., 2023. Extracellular collagenase isolated from Streptomyces antibioticus UFPEDA 3421: purification and biochemical characterization. Prep. Biochem. Biotechnol. 1–12. DOI: 10.1080/10826068.2023.2225090.
- Purification and characterization of a novel extracellular serine-protease with collagenolytic activity from Aspergillus tamarii URM4634. Int. J. Biol. Macromol.. 2018;117:1081-1088.
- [CrossRef] [Google Scholar]
- Purification and characterization of a gelatinolytic serine protease from the seeds of ash gourd Benincasa hispida (Thunb.) Cogn. Indian J. Biochem. Biophys.. 2018;55:77-87.
- [Google Scholar]
- Accurate detection of enzymatic degradation processes of gelatin–alginate microcapsule by 1H NMR spectroscopy: Probing biodegradation mechanism and kinetics. Carbohydr. Polym.. 2023;304:120490
- [CrossRef] [Google Scholar]
- A universal strategy for high-yield production of soluble and functional clostridial collagenases in E. coli. Appl. Microbiol. Biotechnol.. 2009;83:1055-1065.
- [CrossRef] [Google Scholar]
- Applications of MALDI mass spectrometry in clinical chemistry. Clin. Chem.. 2016;62:134-143.
- [CrossRef] [Google Scholar]
- Affinity purification of a recombinant protein expressed as a fusion with the maltose-binding protein (MBP) tag. Methods Enzymol. 2015:17-26.
- [CrossRef] [Google Scholar]
- Dutta, S., Bose, K., 2022. Protein Purification by Affinity Chromatography, in: Textbook on Cloning, Expression and Purification of Recombinant Proteins. Springer Nature Singapore, Singapore, pp. 141–171. DOI: 10.1007/978-981-16-4987-5_6.
- Proteomic protease specificity profiling of clostridial collagenases reveals their intrinsic nature as dedicated degraders of collagen. J. Proteomics. 2014;100:102-114.
- [CrossRef] [Google Scholar]
- Evaluating the utilization of ethylenediaminetetraacetic acid as a treatment supplement for gliomas. Cureus 2022
- [CrossRef] [Google Scholar]
- A novel metalloprotease from banana peel and its biochemical characterization. Int. J. Biol. Macromol.. 2019;134:527-535.
- [CrossRef] [Google Scholar]
- Meng-Chiao Ho, Shih-Hsiung Wu, W.-L.W., 2016. Heat stable keratinase and use thereof. US9434934B2.
- Cloning and characteristic of MMP1 gene from Hyriopsis cumingii and collagen hydrolytic activity of its recombinant protein. Gene. 2019;693:92-100.
- [CrossRef] [Google Scholar]
- Isolation, purification and characterization of collagenase from hepatopancreas of the land snail Achatina fulica. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol.. 2005;142:1-7.
- [CrossRef] [Google Scholar]
- Joseph, J., Sasidharan, H., O.S, C., 2019. Isolation, Production and Characterisation of Novel Gelatinase Enzyme From Bacillus Spp. Int. J. Sci. Res. Biol. Sci. 5, 111–120. DOI: 10.26438/ijsrbs/v5i6.111120.
- Characteristics and applications of a recombinant alkaline serine protease from a novel bacterium Bacillus lehensis. Bioresour. Technol.. 2013;131:76-85.
- [CrossRef] [Google Scholar]
- Purification and biochemical characterization of two novel extracellular keratinases with feather-degradation and hide-dehairing potential. Process Biochem.. 2021;106:137-148.
- [CrossRef] [Google Scholar]
- In vitro Anti-Cancer, Anti-Diabetic, Anti-Inflammation and Wound Healing Properties of Collagen Peptides Derived from Unicorn Leatherjacket (Aluterus Monoceros) at Different Hydrolysis. Turkish J. Fish. Aquat. Sci.. 2019;19
- [CrossRef] [Google Scholar]
- Characterization of chitinase from Shewanella inventionis HE3 with bio-insecticidal effect against granary weevil, Sitophilus granarius Linnaeus (Coleoptera: Curculionidae) Process Biochem.. 2020;97:222-233.
- [CrossRef] [Google Scholar]
- Chitra Latka, Jagga Bikshapathi, Priyanka Aggarwal, Neel Sarovar Bhavesh, R., Chakraborty, S.H.K. and B.T., 2015. Structural analysis of Sub16 sedolisin of Trichophyton rubrum reveals a flexible nature of its pro domain. DOI: DOI: 10.1101/2020.01.28.922815.
- Biotechnological trends in spider and scorpion antivenom development. Toxins (basel).. 2016;8:226.
- [CrossRef] [Google Scholar]
- A novel gelatinase from marine Flocculibacter collagenilyticus SM1988: characterization and potential application in collagen oligopeptide-rich hydrolysate preparation. Mar. Drugs. 2022;20:48.
- [CrossRef] [Google Scholar]
- Antimicrobial and radical scavenging properties of bovine collagen hydrolysates produced by Penicillium aurantiogriseum URM 4622 collagenase. J. Food Sci. Technol.. 2015;52:4459-4466.
- [CrossRef] [Google Scholar]
- Identification of an N-terminal formylated, two-peptide bacteriocin from Enterococcus faecalis 710C. J. Agric. Food Chem.. 2011;59:5602-5608.
- [CrossRef] [Google Scholar]
- In vitro and in silico characterization of alkaline serine protease from Bacillus subtilis D9 recovered from Saudi Arabia. Heliyon. 2021;7:e08148.
- [Google Scholar]
- Nanoformulations based on collagenases loaded into halloysite/Veegum® clay minerals for potential pharmaceutical applications. Colloids Surf., B Biointerfaces. 2023;230:113511
- [CrossRef] [Google Scholar]
- Preparation, characterization, immobilization, and molecular docking analysis of a novel detergent-stable subtilisin-like serine protease from Streptomyces mutabilis strain TN-X30. Int. J. Biol. Macromol.. 2022;222:1326-1342.
- [CrossRef] [Google Scholar]
- Top-down identification and sequence analysis of small membrane proteins using MALDI-MS/MS. J. Am. Soc. Mass Spectrom.. 2022;33:1293-1302.
- [CrossRef] [Google Scholar]
- Comparative analysis of PHAs production by Bacillus megaterium OUAT 016 under submerged and solid-state fermentation. Saudi J. Biol. Sci.. 2020;27:1242-1250.
- [CrossRef] [Google Scholar]
- Antiviral activity of hexapeptides derived from conserved regions of bacterial proteases against HCV NS3 protease. Pak. J. Pharm. Sci.. 2021;34:215-223.
- [CrossRef] [Google Scholar]
- Quantitative analysis of denatured collagen by collagenase digestion and subsequent MALDI-TOF mass spectrometry. Cell Tissue Res.. 2011;343:605-617.
- [CrossRef] [Google Scholar]
- Production, purification, and characterization of Bacillibactin siderophore of Bacillus subtilis and Its application for improvement in plant growth and oil content in sesame. Sustainability. 2021;13:5394.
- [CrossRef] [Google Scholar]
- Characterization and antioxidant activity of collagen, gelatin, and the derived peptides from yellowfin tuna (Thunnus albacares) Skin. Mar. Drugs. 2020;18:98.
- [CrossRef] [Google Scholar]
- OLIVEIRA, V. de M., ASSIS, C.R.D., HERCULANO, P.N., CAVALCANTI, M.T.H., BEZERRA, R. de S., PORTO, A.L.F., 2017. Colagenase de pescada branca: extração, purificação parcial, caracterização e teste de especificidade ao colágeno para aplicação industrial. Bol. do Inst. Pesca 43, 52–64. DOI: 10.20950/1678-2305.2017v43n1p52.
- Identification and homology modeling of a new biotechnologically compatible serine alkaline protease from moderately halotolerant Gracilibacillus boraciitolerans strain LO15. Int. J. Biol. Macromol.. 2020;161:1456-1469.
- [CrossRef] [Google Scholar]
- Microbial collagenases: challenges and prospects in production and potential applications in food and nutrition. RSC Adv.. 2016;6:33763-33780.
- [CrossRef] [Google Scholar]
- Leonard K. Pattenden and Walter G. Thomas, 2008. Affinity Chromatography (Amylose Affinity Chromatography of Maltose-Binding Protein), in: Zachariou, M. (Ed.), Affinity Chromato. Humana Press, Totowa, NJ, pp. 169–190. DOI: 10.1007/978-1-59745-582-4.
- Production and characterization of collagenase from a new Amazonian Bacillus cereus strain. Prep. Biochem. Biotechnol.. 2019;49:501-509.
- [CrossRef] [Google Scholar]
- Discovery and characterization of a new cold-active protease from an extremophilic bacterium via comparative genome analysis and in vitro expression. Front. Microbiol.. 2020;11
- [CrossRef] [Google Scholar]
- Biochemical and molecular characterization of novel keratinolytic protease from Bacillus licheniformis (KRLr1) Front. Microbiol.. 2023;14
- [CrossRef] [Google Scholar]
- Elucidation of structural and functional characteristics of collagenase from Pseudomonas aeruginosa. Process Biochem.. 2018;64:116-123.
- [CrossRef] [Google Scholar]
- The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res.. 2018;46:D624-D632.
- [CrossRef] [Google Scholar]
- Gene cloning, expression, molecular modeling and docking study of the protease SAPRH from Bacillus safensis strain RH12. Int. J. Biol. Macromol.. 2019;125:876-891.
- [CrossRef] [Google Scholar]
- In-vitro and computational analysis of urolithin-A for anti-inflammatory activity on cyclooxygenase 2 (COX-2) Saudi J. Biol. Sci.. 2023;30:103804
- [CrossRef] [Google Scholar]
- Affinity chromatography: a review of trends and developments over the past 50 years. J. Chromatogr. B. 2020;1157:122332
- [CrossRef] [Google Scholar]
- MALDI-TOF MS and CD spectral analysis for identification and structure prediction of a purified, novel, organic solvent stable, fibrinolytic metalloprotease from Bacillus cereus B80. Biomed Res. Int.. 2015;2015:1-13.
- [CrossRef] [Google Scholar]
- Processing of protease under sub- and supercritical conditions for activity and stability enhancement. Biochem. Eng. J.. 2014;92:83-89.
- [CrossRef] [Google Scholar]
- Biochemical, thermodynamic and structural characteristics of a biotechnologically compatible alkaline protease from a haloalkaliphilic, Nocardiopsis dassonvillei OK-18. Int. J. Biol. Macromol.. 2020;153:680-696.
- [CrossRef] [Google Scholar]
- Gene expression and molecular characterization of recombinant subtilisin from Bacillus subtilis with antibacterial, antioxidant and anticancer properties. Int. J. Biol. Macromol.. 2023;249:125960
- [CrossRef] [Google Scholar]
- Jinsong Shi, Zhenghong Xu, Jinsong Gong, Heng Li, C.S., 2021. Industrial Keratinase via genetic enginering and use thereof. US11041160B2.
- Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol.. 2011;7
- [CrossRef] [Google Scholar]
- A microscale enzyme experiment based on bacterial gelatinase. Acta Manila. Ser. a.. 2015;63:97-102.
- [CrossRef] [Google Scholar]
- Naringin dihydrochalcone potentially binds to catalytic domain of matrix metalloproteinase-2: molecular docking, MM-GBSA, and molecular dynamics simulation approach. Nat. Prod. Res.. 2023;37:1851-1855.
- [CrossRef] [Google Scholar]
- Identification and characterization of a protease (EuRP-61) from Euphorbia resinifera latex. Int. J. Biol. Macromol.. 2020;145:998-1007.
- [CrossRef] [Google Scholar]
- Identification and antioxidant activity of bovine bone collagen-derived novel peptides prepared by recombinant collagenase from Bacillus cereus. Food Chem.. 2021;349:129143
- [CrossRef] [Google Scholar]
- Characterisation of proteolytic enzymes from muscle and hepatopancreas of fresh water prawn (Macrobrachium rosenbergii) J. Sci. Food Agric.. 2011;91:52-59.
- [CrossRef] [Google Scholar]
- A combination of bioinformatics analysis and rational design strategies to enhance keratinase thermostability for efficient biodegradation of feathers. Sci. Total Environ.. 2022;818:151824
- [CrossRef] [Google Scholar]
- Cloning, expression, purification and characterisation of serine alkaline protease from Bacillus subtilis RD7. Biocatal. Agric. Biotechnol.. 2019;20:101264
- [CrossRef] [Google Scholar]
- Extraction, partial purification and characterization of alkaline protease from rainbow trout (Oncorhynchus Mykiss) viscera. Aquaculture. 2019;500:458-463.
- [CrossRef] [Google Scholar]
- Recombinant collagenase from Grimontia hollisae as a tissue dissociation enzyme for isolating primary cells. Sci. Rep.. 2020;10:3927.
- [CrossRef] [Google Scholar]
- Cloning of a Novel Collagenase Gene from the Gram-Negative Bacterium Grimontia (Vibrio) hollisae 1706B and Its Efficient Expression in Brevibacillus choshinensis. J. Bacteriol.. 2011;193:3049-3056.
- [CrossRef] [Google Scholar]
- Heterologous Expression and Structural Elucidation of a Highly Thermostable Alkaline Serine Protease from Haloalkaliphilic Actinobacterium, Nocardiopsis sp. Mit-7. Appl. Biochem. Biotechnol. 2023
- [CrossRef] [Google Scholar]
- Screening collagenase activity in bacterial lysate for directed enzyme applications. Int. J. Mol. Sci.. 2021;22:8552.
- [CrossRef] [Google Scholar]
- Application and perspectives of MALDI–TOF mass spectrometry in clinical microbiology laboratories. Microorganisms. 2021;9:1539.
- [CrossRef] [Google Scholar]
- Isolation and Characteristics of Bacillus subtilis CN2 and its collagenase production. J. Food Sci.. 2002;67:1184-1187.
- [CrossRef] [Google Scholar]
- Characterization of bovine neutrophil gelatinase-associated lipocalin. J. Dairy Sci.. 2006;89:3400-3407.
- [CrossRef] [Google Scholar]
- Wanderley, M.C. de A., Duarte Neto, J.M.W., Andrade, A.F. de, Melo, R.G. de, Viana-Marques, D. de A., Bezerra, R.P., Porto, A.L.F., 2020. First report on Chlorella vulgaris collagenase production and purification by aqueous two-phase system. Sustain. Chem. Pharm. 15, 100202. DOI: 10.1016/j.scp.2019.100202.
- Mechanistic Insight into the fragmentation of type I collagen fibers into peptides and amino acids by a vibrio collagenase. Appl. Environ. Microbiol.. 2022;88
- [CrossRef] [Google Scholar]
- Purification and Characterization of a Novel Collagenase from Bacillus pumilus Col-J. Appl. Biochem. Biotechnol.. 2010;160:129-139.
- [CrossRef] [Google Scholar]
- Optimization of collagenase production by Pseudoalteromonas sp. SJN2 and application of collagenases in the preparation of antioxidative hydrolysates. Mar. Drugs. 2017;15:377.
- [CrossRef] [Google Scholar]
- Genome analysis of Bacillus aryabhattai to identify biosynthetic gene clusters and in silico methods to elucidate its antimicrobial nature. Int. J. Pept. Res. Ther.. 2021;27:1331-1342.
- [CrossRef] [Google Scholar]
- In Silico Study on the Inhibition of UDP-N-Acetylglucosamine 1-Carboxy Vinyl Transferase from Salmonella typhimurium by the Lipopeptide Produced from Bacillus aryabhattai. Int. J. Pept. Res. Ther.. 2022;28:80.
- [CrossRef] [Google Scholar]
- Gene Expression and Characterization of Iturin A Lipopeptide Biosurfactant from Bacillus aryabhattai for Enhanced Oil Recovery. Gels. 2022;8:403.
- [CrossRef] [Google Scholar]
- Gelatin Extracted from Rahu (Labeo rohita) and Silver Carp (Hypophthalmichthys molitrix) Scales and their Beads Efficacy as a Carrier of Inoculum and Secondary Metabolites. Biosci. Rev.. 2022;4:59-72.
- [CrossRef] [Google Scholar]
- Li-Jung Yin, Ya-Hui Chou, and S.-T.J., 2013. PURIFICATION AND CHARACTERIZATION OF ACIDIC PROTEASE FROM ASPERGILLUS ORYZAE BCRC 30118. J. Mar. Sci. Technol. Vol. 21, N, 105–110. DOI: 10.6119/JMST-012-0529-1.
- Probing the Crucial Role of Leu31 and Thr33 of the Bacillus pumilus CBS Alkaline Protease in Substrate Recognition and Enzymatic Depilation of Animal Hide. PLoS One. 2014;9:e108367.
- [Google Scholar]
- Refolding of recombinant histidine-tagged catalytic domain of MMP-13 from Escherichia coli with ion-exchange chromatography for higher bioactivity. J. Liq. Chromatogr. Relat. Technol.. 2015;38:561-568.
- [CrossRef] [Google Scholar]
- Optimized recombinant expression and characterization of collagenase in Bacillus subtilis WB600. Fermentation. 2022;8:449.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2024.105942.
Appendix A
Supplementary data
The following are the Supplementary data to this article:Supplementary Data 1
Supplementary Data 1







