Translate this page into:
New superior bioactive metal complexes of ligand with N, O donor atoms bearing sulfadiazine moiety: Physicochemical study and thermal behavior for chemotherapeutic application
⁎Corresponding author. sasa.memo27@gmail.com (Sabreen Mohamed El-Gamasy)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Sulfadiazine is a drug famous for its anti-inflammatory, antimicrobial, and antitumor effects. Remarkably, its biological activity can be further enhanced upon incorporating a suitable metal ion. However, these metal-comprising complexes are not widely available. In the current study, a sulfadiazine Schiff base derivative was exploited as a ligand for synthesizing new complexes utilizing Cu(II), Co(II), Ni(II), Fe(III), and Cr(III) metals. The structural and analytical characteristics of the ligand and the newly prepared complexes were elucidated using various spectral and thermal investigations. Also, the biological activity of the ligand and the metal complexes, including the cytotoxic effect on normal cells and on liver malignant cells and the antimicrobial activity, was examined. The infrared spectra demonstrated that the ligand coordinated to all the added metal ions in a neutral form. It behaved in a bidentate manner in all mononuclear complexes. The new complexes exhibited octahedral geometry. Evaluating the biological activities revealed that the ligand and its novel metal-containing complexes had moderate antimicrobial activity, while the metal complexes, especially those comprising of Cr(III), Fe(III), and Cu(II), displayed a superior chemotherapeutic effect on HepG2 cell line in comparison to the ligand with very week or rare cytotoxic effects on normal human cells. Efficiently, new sulfadiazine Schiff base derivative-containing metal complexes with enhanced therapeutic potential were manufactured and could be applied on experimental models for the treatment of various types of infections and malignancies.
Keywords
Antimicrobial activity
Chemotherapy
Sulfadiazine derivative
Thermal analysis
1 Introduction
Sulfa containing drugs are chemotherapeutic agents that consist of molecular structures containing 4-amino benzene sulfonamide moiety (Radha et al., 2016). In addition to the low price and the low toxic effect, sulfonamides have considerable antiviral, antifungal, anti-inflammatory, and antitumor activities which make them among the most broadly used therapeutic drugs (Thangavelu and Thangavel, 2018). Sulfadiazine, a sulphanilamide derivative, is well-known as an antibacterial, as well as, an antimalarial drug (Modhavadiya, 2017). It is thought that its biological activity is derived from the structural semblance between the sulfanilamide group and the para-aminobenzoic acid which is a competitive inhibitor, inhibiting completely the action of dihydropteroate synthase, and thus, blocking the folic acid synthesis pathway and cell division, causing cell death (Jain and Singh, 2006) (see Scheme 1).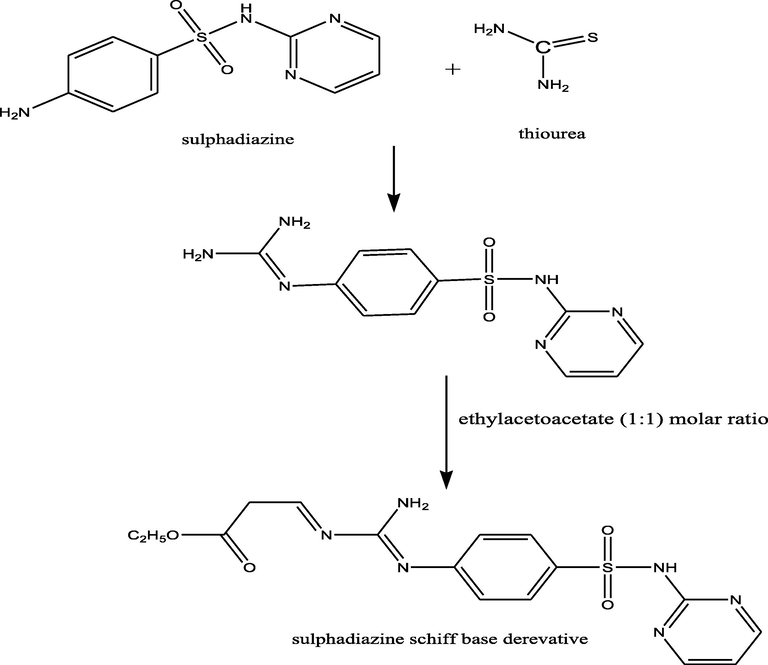
Synthesis of the ligand (1).
Due to the global impact of cancer diseases, several strategies have emerged to augment the biological and anticancer activities of the available medications by modifying their chemical assembly (Thangavelu and Thangavel, 2018). Studies, carried out to innovate new metal-based drugs, are both promising and of great value for the development of more effective therapeutic agents (Ndagi et al., 2017). Compounds containing sulfonamide group have long been used as a treatment for many medical infections and disorders. It has been stated that the activity of many of these complexes can be increased upon incorporating a suitable metal ion (Maurya et al., 2016; Govindaraj and Ramanathan, 2014). The efficient development of metal complexes of sulfadiazine has recently resumed the worth of these compounds in medicine (Ajibade and Okoh, 2013). Currently, sulfadiazine-containing compounds are commonly used to block bacterial infection associated with burn treatment. Also, the investigation of metal-based sulfonamides was motivated by the fruitful production of Ag(I) and Zn(II) sulphadiazine complexes to prevent various bacterial infections (Chohan and Naseer, 2007). Fortunately, these metal complexes are widely available. This consideration provoked us to synthesize a new series of metal-associated sulfadiazine complexes and investigate their anticancer and antibacterial activities.
Consequently, this study was aiming to synthesize, to characterize, and to examine the biological and therapeutic activities of sulfadiazine drug derivative and its transition metal-based complexes which attract much attention because of their interesting anti-microbial and anti-tumor properties.
2 Experimental
2.1 Physical measurements
A standard gravimetric method was applied to reveal the metal ion (Vogel, 1966). All metal-comprising complexes were dried out under vacuum over P4O10. The infrared (IR) spectra were analyzed as KBr and CeBr pellets utilizing Perkin-Elmer 683 spectrophotometer (4000–200 cm−1). The electronic spectral data were qualitatively obtained by Perkin-Elmer 550 spectrophotometer. The conductance of metal complexes (1 × 10−3 M solution in DMF) was assessed, at room temperature, using Bibby conductimeter. The 1H NMR spectra of both the ligand and the Cu(II) and Zn(II) complexes were measured by Perkin-Elmer R32-90-MHz spectrophotometer with TMS being utilized as an internal standard. The mass spectrum of the ligand was determined using JEULJMS-AX-500 mass spectrometer connected to data software. Thermal analyses (DTA and TGA) were done with a temperature ranging from 27 to 800 °C, at a heating rate of 10 °C/min, utilizing Shimadzu DT-30 thermal analyzer. Gouy’s method was applied to measure the magnetic susceptibilities using mercury tetrathiocyanate cobalt(II) as a standard while Pascal constant (Lewis et al., 1967) was used to evaluate the diamagnetic corrections. The magnetic moments were calculated according to equation. The ESR spectra of the metal complexes were identified by Varian E-109 spectrophotometer, at room temperature, with DPPH used as a standard. The TLC verified the purity of all compounds.
2.2 Synthesis of the ligand and its metal complexes
2.2.1 Synthesis of the ligand, [L], (1)
Ligand 1 was synthesized by refluxing sulfadiazine drug [4-amino-N-pyrimidin-2-yl-benzenesulfonamide] (30 g, 0.1191 mol) and thiourea (9.124 g, 0.1191 mol). Then, ethyl acetoacetate (15.493 g, 0.1191 mol) was added to the product at a molar ratio of 1:1 in 100 ml ethanol with stirring for 4 h. The product was left to cool at room temperature, and then it was filtered to separate the sulfadiazine Schiff base. The precipitate was filtered and washed by ethanol several times. Finally, it was dried over P4O10 in vacuum.
2.2.2 Synthesis of metal complexes (2)–(11)
Ligand (1) (5.0 g, 0.0124 mol), prepared in 50 ml ethanol, was added to 30 ml an ethanolic solution of (2.46 g, 0.0124 mol) of Cu(OAc)2·H2O, generating complex (2), (2.94 g, 0.0124 mol) of Cu(NO3)2·.5H2O, to prepare complex (3), (2.11 g, 0.0124 mol) of CuCl2·2H2O synthesizing complex (4), (3.09 g, 0.0124 mol) of CuSO4·5H2O, to form complex (5), (3.09 g, 0.0124 mol) Co(OAc)2·.4H2O, to generate complex (6), (3.08 g, 0.0124 mol) Ni(OAc)2·4H2O, producing complex (7), (3.45 g, 0.0124 mol) of FeSO4·7H2O, to make complex (8), (4.85 g, 0.0124 mol) of Cr2(SO4)3.6(H2O), forming complex (9), (3.25 g, 0.0124 mol) of NiSO4·6H2O, to develop complex (10), (2.72 g, 0.0124 mol) of Zn(OAc)2·2H2O, to prepare complex (11). The ratio of the ligand to metal, in the complexes, was 1:1. The mixtures were stirred, for 6–8 h, dependent on the character of the metal ion and the anion. When a precipitate was noticed, it was filtered, washed with ethanol, and dried over P4O10 in vacuum.
2.3 Cytotoxic effect of ligand and complexes on human cells
Fifteen milliliters of blood was withdrawn from a participant after signing informed consent. The blood (15 ml) was carefully layered over Bicoll in a 15 ml conical tube at a ratio of 1.5 to 1. Centrifugation was carried out at 2000 rpm for 25 min at 20 °C. The mononuclear cell layer was collected and washed twice with phosphate buffer saline (PBS). The supernatant was discarded and the pellet was dislodged. Red blood cells (RBCs) were lysed by lysing buffer followed by centrifugation then the pellet was washed with PBS (Soliman et al., 2017). The cell pellet was resuspended in complete Dulbecco’s Modified Eagle’s Medium (DMEM) which consisted of low-glucose DMEM (Gibco, USA), 30% fetal bovine serum (Biochrom, Germany), 1% penicillin/streptomycin (100 U/mL), and 1% l-glutamine. Cell count and viability were calculated by hemocytometer. The mononuclear cells were cultured in 96-well plates containing complete DMEM, at a seeding density of 1 × 104 cells/well, and the ligand and complexes were added to the cells (1000 µg/mL). Serial dilution of the complexes was done 5 times until reaching a concentration of 31.2 µg/mL. Finally, the cells were incubated in 5% CO2 and 90% humidified atmosphere at 37 °C for 24 h, and then, cells were counted visually under an inverted microscope (Abd El-RazekZ et al., 2020). Based on the United States National Cancer Institute categorization, the activity of cytotoxicity was classified as high activity when IC50 was below 20 μg/mL, moderate activity when IC50 was 20–100 μg/mL, weak activity when IC50 was 201–500 μg/mL, and no activity when IC50 exceeded 500 μg/mL (Huda et al., 2017).
2.4 Chemotherapeutic effect
The chemotherapeutic effect was evaluated based on the cytotoxic activity of the complexes on human hepatoma cells (HepG2). HepG2 cell line passage number 80≈85 was separately cultured in 25 cm2 tissue culture flasks using RPMI medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin. Afterwards, the flasks were incubated in CO2 incubator at 37 °C until 90% confluence was reached. Then, the media were carefully removed and the flasks were washed with PBS once. Trypsin (0.25%) was added for 7 min and the flasks were checked constantly under an inverted microscope until all the cells were detached. Subsequently, the collected cells were washed with PBS, resuspended in culture media, and inoculated, at a density of 104 cells/well, in 96-well plates which were incubated until sheets of cells were observed. Serial dilutions of drug complexes, in growth media, were added to cells at dilutions of 1000, 500, 250, 125, 62.5, and 31.2 µg/mL, followed by, incubation of the plates for 24 h. The malignant cell viability was assessed by crystal violet staining where crystal violet stain was utilized to stain DNA and to determine the density of well-adherent viable cells. Initially, the media were decanted, the plates were washed with PBS, the cells were stained with 20 µl of 0.5% crystal violet (Sigma-Aldrich Corp., USA), in 30% ethanol, and were left at room temperature for 10 min. Then, the plate wells were cautiously washed 3 times with distilled water. Lastly, the color absorbance values were obtained at an optical density (OD) of 490 nm (Śliwka et al., 2016).
2.5 Antimicrobial activity
The cup-plate agar diffusion approach was done to assess the antimicrobial effect of the different chemical complexes dissolved in DMSO (200 µg/100 µl) (Valgas et al., 2007). Two bacterial microbes; Staphylococcus aureus ATCC6538-P (Gram +ve) and Pseudomonas aeruginosa ATCC 27853 (Gram −ve); one yeast test microbe Candida albicans ATCC 10231, and one fungal test microbe Aspergillus niger NRRL A-326 were picked to determine the antimicrobial activities of the ligand and its complexes. The bacterial and the yeast microbes were grown on nutrient agar medium (g/L): agar (20), beef extract (3), and peptone (10). Whereas, the fungal test microbe was grown on Szapek-Dox agar medium (g/l): agar (20), Sucrose (30), NaNO3 (3), MgSO4·7H2O (0.5), KCl (0.5), FeSO4·7H2O (0.055), and K2HPO4 (1). The culture media of each test microbe was diluted with pre-sterilized distilled water to 107–108 colony forming units (CFU)/mL. One mL of each was inoculated into 1 L-Erlenmeyer flask having 250 ml of solidified agar media (Youssef et al., 2014). 25 ml of these media were poured in sterilized Petri dishes (10 cm diameter). In each plate, 3–4 cups, measuring 10 mm in diameter were cut. The cups were then filled with 100 uL of samples which were allowed to diffuse, for 2 h, at 4 °C. Afterwards, the inoculated plates were incubated, for 24 h, at 37 °C (for bacteria) and for 48 h, at 30 °C (for fungus). Finally, the diameters of the microbial growth inhibition zones, resulting from the ligand and metal complexes, were measured.
3 Results and discussion
All analytical, physical (Table 1), and spectral data (Tables 2 and 3) showed that the newly synthesized compounds were produced at a ligand to metal stoichiometric ratio of 1:1. All the complexes were insoluble in common solvents, viz: MeOH, EtOH, CHCl3, CCl4, and (CH3)2CO but soluble in DMSO. Ligand 1 was prepared by the condensation of sulfadiazine drug with thiourea at a molar ratio of 1:1. Then, ethyl acetoacetate was added to the product at a molar ratio of 1:1 as shown in Fig. 1.
No.
Ligand/Complexes
Color
F.W.
M.P. (°C)
Yield (%)
Anal./Found (Calc.) (%)
Molar conductance Λm (Ω−1 cm2 mol−1)
C
H
N
M
Cl
(1)
[L]
C17H20N6O4SYellow
404.43
280
80
50.99
(50.49)5.02 (4.95)
20.89 (20.79)
–
–
–
(2)
[(L)Cu(OAc)2(H2O)2]
C21H30CuN6O10SGreen
621.5
>300
70
40.33
(40.55)4.9 (4.83)
13.66 (13.52)
10.41 (10.22)
–
15
(3)
[(L)Cu(SO4)(H2O)3].2H2O
C17H30CuN6O13S2
Dark green
653.5
290
76
31.74
(31.22)4.64 (4.59)
12.77 (12.85)
9.79
(9.72)–
17
(4)
[(L)CuCl2(H2O)2].H2O
C17H26Cl2CuN6O7SPale green
592.5
295
78
34.63
(34.43)4.53 (4.39)
14.38 (14.18)
10.80 (10.72)
11.98
(11.88)14
(5)
[(L)Cu(NO3)2(H2O)2]. 2H2O
C17H28CuN8O14SGreen
663.5
>300
78
30.87
(30.74)4.33 (4.22)
16.99 (16.88)
9.80
(9.57)–
15
(6)
[(L)Co(OAc)2(H2O)2]. 2H2O
C21H34CoN6O12SBrown
652.9
>300
76
38.44
(38.59)5.28 (5.21)
12.70 (12.87)
9.09
(9.02)–
13
(7)
[(L)Ni(OAc)2(H2O)2].3H2O
C21H36NiN6O13SGreen
618.6
>300
70
40.76
(40.74)5.77 (5.82)
13.54 (13.58)
9.50
(9.47)–
12
(8)
[(L)Fe(SO4)(H2O)3]. 2H2O
C17H30FeN6O13S2
Brown
645.8
>300
79
31.89
(31.59)4.57
(4.65)13.08
(13.00)8.60
(8.64)–
177
(9)
[(L) Cr(SO4)(H2O)3]. 3H2O
C17H32CrN6O14S2
Dark green
659.9
>300
69
30.94
(30.91)4.79 (4.85)
12.74 (12.73)
7.78
(7.86)–
167
(10)
[(L)Ni(SO4)(H2O)3].2H2O
C17H30NiN6O13S2
Green
648.6
>300
78
31.61
(31.45)4.73 (4.63)
12.87 (12.95)
9.08
(9.03)–
11
(11)
[(L) Zn(OAc)2(H2O)2].H2O
C21H38ZnN6O14SPale yellow
695.39
>300
78
36.31
(36.24)5.69 (5.64)
12.04 (12.08)
9.37
(9.4)–
17
No.
ν(H2O/NH2)
ν(H-bond.)
ν (NH)
ν(C⚌N)
ν(C⚌O)
ν(S—N)
ν(SO2 - N) moiety
ν(Ar)
υ (OAc)/SO4/[NO3]
υ(M—O)
υ(M—N)
υ(M—Cl)
(1)
3450
3430–3210
3200–30503560–3070
3000–25403200
1620
1665975
1350, 11551605, 752
–
–
–
(2)
3420
3550–3310
3300–30203580–3280
3302–23403254
1610
1654965
1320, 11251604, 736
(1530, 1390)
520
465–
(3)
3395
3560–3220
3200–30103610–3280
3270–27603230
1609
1645960
1310, 1120
1575, 740
1105, 1030, 805, 690, 485
530
440–
(4)
3350
3630–3320
3290–30403580–3260
3250–29003270
1603
1670940
1315, 1110
1595, 745
–
535
465425
(5)
3410
3616–3190
3140–30203630–3210
3190–28603220
1612
1655945
1325, 1128
1563, 730
1385, 1285, 815, 765
560
485–
(6)
3387
3780–3210
3160–30203620–3160
3070–27003225
1605
1660950
1330, 11401562, 740
(1515, 1385)
556
430–
(7)
3365
3620–3310
3190–28003640–3220
3160–28003250
1607
1670960
1335, 1135
1565, 737
(1520, 1360)
530
460–
(8)
3360
3615–3260
3180–30503654–3180
3115–25603265
1606
1662955
1329, 11301590, 734
1108, 1030,
820, 640, 400539
470–
(9)
3355
3605–3370
3150–3050
3640–3100
3180–27603240
1608
1654935
1335, 11401558, 740
1115, 1020, 805, 625, 420
545
448–
(10)
3370
3510–3250
3230–30603620–3150
3120–28003260
1604
1663930
1315, 11151560, 760
1108, 1010,
802, 620,
410536
455–
(11)
3355
3645–3255
3200–30603650–3200
3270–25003280
1602
1667955
1322, 11091575, 745
(1510, 1410)
540
465–
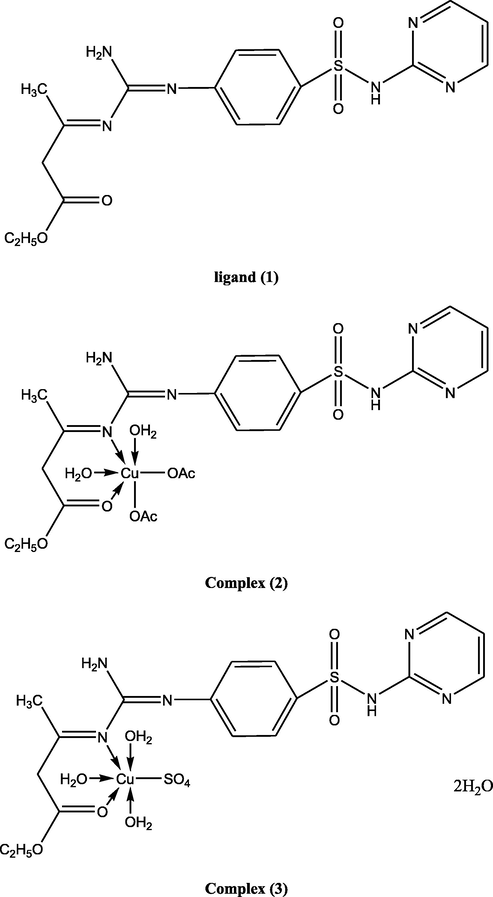
Suggested structures of ligand and its metal complexes.
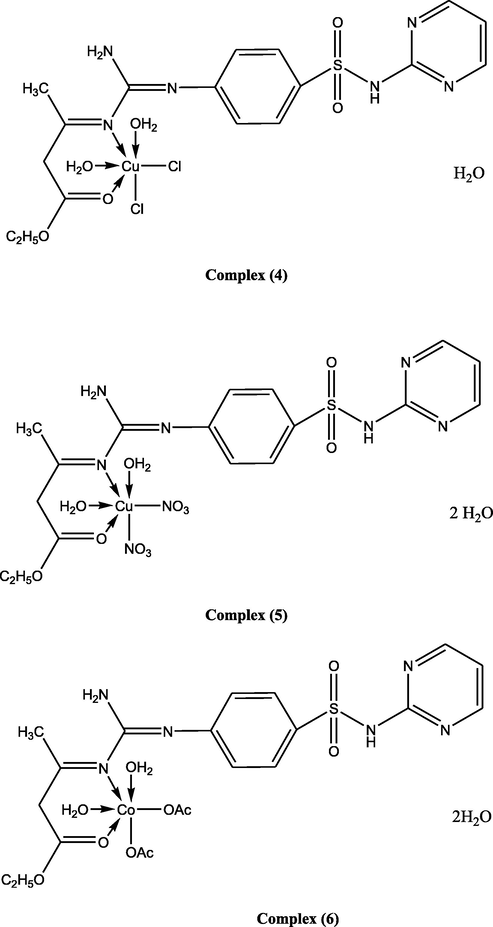
Suggested structures of ligand and its metal complexes.
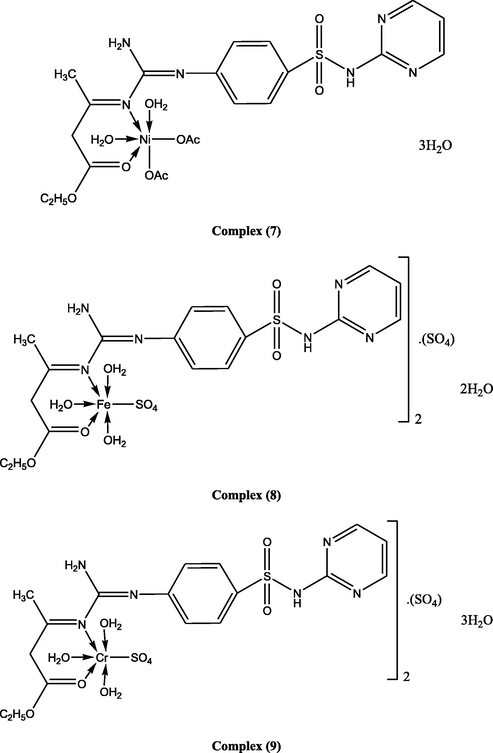
Suggested structures of ligand and its metal complexes.
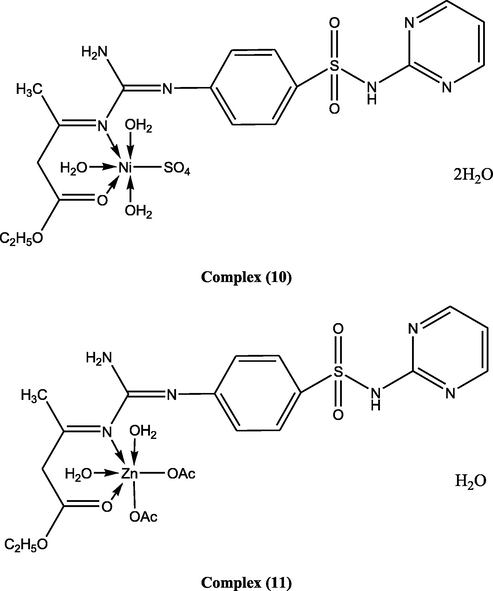
Suggested structures of ligand and its metal complexes.
3.1 Mass spectrum of the ligand, (1) and its complexes (2, 4)
The mass spectrum of the ligand revealed a molecular ion peak (m/z) at 404.43, which was coincident with its calculated molecular weight. Proposed mass fragmentation of ligand 1 was illustrated in Fig. 2.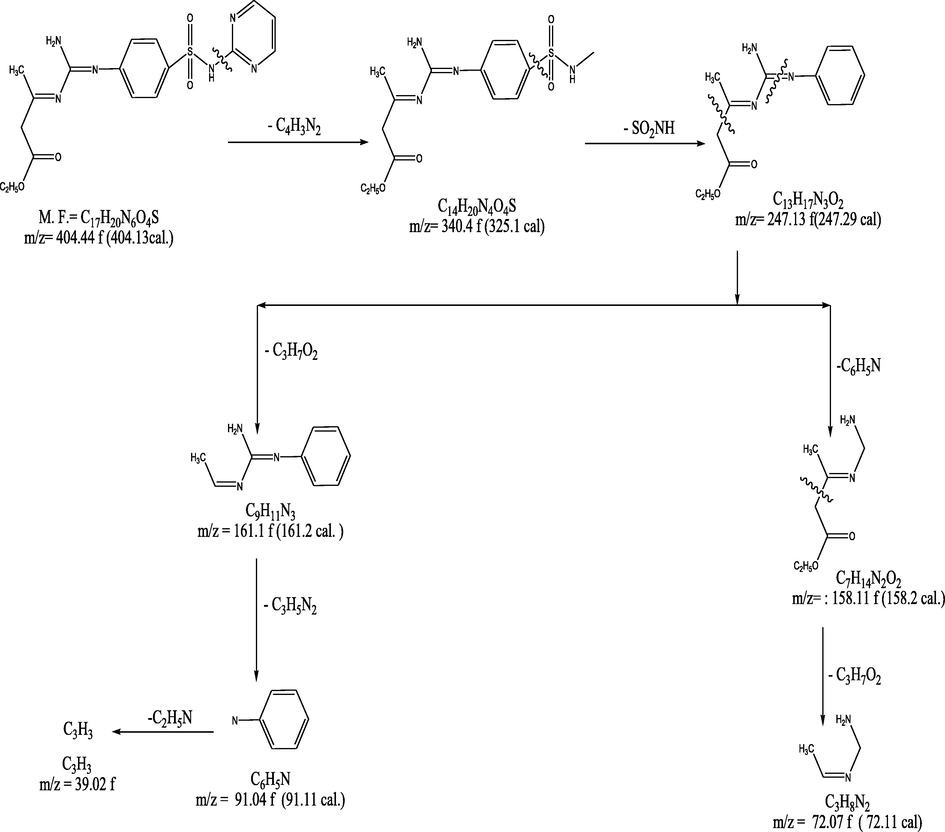
Proposed mass fragmentation of ligand (1).
The mass spectrum of Cu(II)-containing complex (2) indicated a molecular ion peak at 621.5 m/z, coincident with its molecular weight, and the fragmentation pattern split a parent ion peak at (m/z) = 79, 94, 158, 234, 276, 387.5, 523.5, 576.5, and 621.5 a.m.u. corresponding to C4H3N2, C4H4N3, C4H4 N3SO2, C10H8N3SO2, C11H10N5SO2, C11H12N6SO4Cu, C15H20N6 SO9Cu, C19H25N6SO9Cu, and C21H30N6SO10Cu moieties, respectively. The mass spectrum of Cu(II)-comprising complex (4) demonstrated a molecular ion peak at 592.5 a.m.u. coincident with its molecular weight and the fragmentation pattern split a parent ion peak (m/z) at 18, 97, 112, 176, 252, 294, 460.5, 494.5, 547.5, and 592.5 a.m.u. corresponding to H2O, C4H5N2O, C4H6N3O, C4H6N3SO3, C10H10N3SO3, C11H12N5SO3, C13H14N6SO4Cl2Cu, C11H16N6SO6Cl2Cu, C15H21N6SO6Cl2Cu, and C17H26N6SO7Cl2Cu moieties, respectively (supporting information table 2).
3.2 1H – NMR spectra
The 1H – NMR spectra of the ligand and its Zn(II) metal complex (11), in deuterated DMSO, identified signals consistent with the structure. The NMR showed the differences in the magnetic properties of the various nuclei present and deduced in a large measure where these atoms are within the molecule. The NMR studies also detected the change in the chemical shift of the complexes and the ligand as a result of complexation. The signals observed, at 4.15 and at 3.25 ppm, were owing to the protons of two NH2 groups (Jeffrey et al., 2015). The proton of azomethine group appeared at 10.4 ppm. The peaks were identified as multiple ones at 6.1–7.5 ppm may be assigned to aromatic protons (Mayadevi et al., 2003). The protons in the ligand were shifted downfield because of the metal ions (Balgavý et al., 1980).
The NMR spectral data of Zn(II) complex (11) revealed changes in the chemical shift and intensity of the bands in relation to those of the free ligand which indicated the addition of the metal to the free ligand proving that the azomethine nitrogen atom coordinated to the metal ion (N - Zn) (Kavitha and Anantha Lakshmi, 2017). Upon complexation, the sulfa-H imino proton signal disappeared suggesting the covalent binding of sulfonamide nitrogen to the metal atom. The aromatic protons appeared in the 6.3–7.9 ppm range. A new signal was noticed at 1.87 ppm produced by the coordinated acetate group protons (Kavitha and Anantha Lakshmi, 2017).
3.3 Conductivity
The molar conductance measurements of the prepared compounds in DMF (1 × 10−3 M) presented in (Table 1) indicating that the complexes (2–7), (10), and (11) were not electrolytes. This confirmed that the anion was incorporated into the metal ion. On the other side, complexes (8) and (9) showed a different property as their conductivity values were 165 and 177 Ω mol−1cm2, respectively, which might be caused by the expanding ionization potential of the central metal ions that enhanced the lattice energy. The molar conductivity findings showed that the complexes were electrolytes in the solvent DMF and established the stability of theses complexes.
3.4 IR spectra
Careful investigation of the IR spectra of complexes and comparison with that of the ligand was carried out giving information to illustrate the manner of bonding of the ligand towards different metal ions. Thus a detailed characterization of the IR spectrum of the ligand and the effect of metal ion binding on the vibration frequencies was performed. The most fundamental IR spectral bands of all systems and their assignments are collected in (Table 2). The data elicit the following observations:
-
The enolization of sulfonamide moiety in sulfadiazine Schiff base ligand (1) predicated to S⚌O and NH groups, where one NH is responsible for enol – keto form and the other has the same nature as free ligand, as indicative of different mode of vibrations of NH group (ν, δ, γ) (Kavitha and Anantha Lakshmi, 2017).
-
All complexes showed the same modes of vibrations corresponding to NH and S⚌O groups, indicating that the ligand chelates with metal ions in neutral keto–enol–form. The (ν, δ, γ) of NH group showed that sulfonamide moiety doesn’t participate in metal complexation with different metal ions (Kavitha and Anantha Lakshmi, 2017).
-
All complexes display ν(C⚌N) and ν(C⚌O) bands that are shifted to higher wavenumbers than those of free ligand by 2–18 and 3–20 cm−1, respectively, suggesting the participation of nitrogen atom of azomethene group in bonding and oxygen of carbonyl group. (Fouda et al., 2008; Youssef et al., 2010)
-
The ν(SO2-NH) group and ν(NH2) ligand bands showed weak effect for all complexes indicating that sulfonamide moiety atoms of sulfadiazine and nitrogen of amino group are not involved in bonding. (Kavitha and Anantha Lakshmi, 2017; Youssef et al., 2010)
-
The spectra of the complexes gave new bands, not observed in that of the ligand, at 560–520 and 470–430 cm−1 assignable to ν(M—N) and ν(M—O). (Youssef et al., 2010; El-Tabl et al., 2015) The ν(C⚌O) predicated to C⚌O in all complexes are coordinated with different metal ions.
-
Additional to the above observation, the ν(NH2) group band shows more or less shift around 3450 cm−1 and a change in its shape. This is may be due to the formation of inter‐ or intramolecular hydrogen bonding. According to the above observations, the ligand behaves as a neutral bidentate ligand and binds with different metal ions through nitrogen and oxygen atoms of azomethene and carbonyl groups complexes, ν(C⚌N) and ν(C⚌O) bands are shifted to higher and lower wavenumber by 2–18 and 3–20 cm−1 associated to emphasize the coordination of azo‐nitrogen atom and oxygen of carbonyl group with the metal ions.
-
The ligand behaves as a bidentate moiety and chelates metal ions through both carbonyl oxygen atoms of ligand and with azo nitrogen atom.
-
In complexes (2), (6), (7), and (11) the bands were produced by νa(COO) noticed in the 1530–1510 cm−1 and the νs(COO) observed in the 1410–1360 cm−1 ranges. The disparity between these two bands was in the 20–50 cm−1 range signifying that the acetate group coordinated in a unidentate manner with the metal ions (Fouda et al., 2008). Complex (5) showed bands at (1385, 1285, 815, 765) cm−1 which were assigned to coordinated nitrate group (Fouda et al., 2008).
-
Whereas, sulphate complexes (3) and (8–10) exhibited new bands at (1105, 1030, 805, 690, 485), (1108, 1030, 820, 640, 400), (1115, 1020, 805, 625, 420), and (1108, 1010, 802, 620, 410) cm−1 which implied that the sulphate ion coordinated to the metal ion in a unidentate chelating way (El-Tabl et al., 2008; El-Tabl et al., 2013).
-
Complex (4) showed a band at 425 cm−1 which was generated by the coordinated monodentate chloride anion ν(M—Cl) (El-Tabl et al., 2008).
3.5 Electronic transition and magnetic moments
The DMF electronic absorption spectra and the effective magnetic moment values of the ligand and of its metal complexes are reported in Table 4. The ligand displayed three transition bands in the high energy region. The first band which presented at 255 nm was appointed to π → π* transition within the aromatic rings and this band was nearly the same upon complexation. The second and third bands detected at 270 and 330 nm were due to n → π* of the azomethine groups and ν(SO2 - N) moiety and CT transitions (Mohamed et al., 2005; Aslan et al., 2011). These bands were shifted upon complexation providing the involvement of these transitions in the coordination with the new metal ions. The electronic spectra of Cu(II) complexes (2–5) exhibited bands in the 452–630, 455–645, 460–655, and 445–648 nm ranges which were designated to 2B1g→ 2A1g (d(x2−y2) → dz2) and 2B1g→2Eg (d(x2−y2) → dxy, dyz) transitions, respectively which denoted that the Cu(II) ion had a tetragonally distorted octahedral geometry. This could be attributed to the Jahn-Teller effect operated on the d9 electronic ground state of six coordinate system, elongating one trans pair of coordinate bonds and shortening the remaining four ones (Mohamed et al., 2005; Aslan et al., 2011). The magnetic moments of all Cu(II) complexes (2–5) were in the 1.68–1.74B.M. range indicated that the complexes had square planar and octahedral geometry (Kumar et al., 2014). The spectra of Ni(II) complexes (7) and (10) showed three bands at (495, 620, 735), and at (496, 618, 736) nm, these bands were corresponding to 3A2g (F) → 3T2g (F)(1), 3A2g (F) → 3T1g(F)(2), and 3A2g (F) → 3T1g(P)(3) transitions, respectively, entailing octahedral Ni(II) complexes formation (Akbar et al., 2011). The lowest value of 2/1 ratio for the complexes was within 1.27–1.34 range which was below the usual range of 1.5–1.75 which was indicative of distorted octahedral Ni(II) complex (Akbar et al., 2011). The magnetic moment values of Ni(II) complexes (7) and (10) were 2.77 and 2.9B.M., respectively, in harmony with two unpaired electrons state and proving octahedral geometry of the Ni(II) ion (Akbar et al., 2011). The electronic spectra of the Co(II) complex (6) showed three d–d transition bands at (555, 620, 715 nm). These three bands were assigned to 4T1g(F)→4T2g(F)(1), 4T1g(F) →4T1g(p)(2), and 4T1g(F) →4A1(F)(3) transitions, respectively, consistent with high spin Co(II) octahedral complexes (Mohamed et al., 2005). The magnetic moment of complex (6) was 4.7B.M. which was within the described range of high spin octahedral Co(II) complex. Iron(III) complex (8) showed six bands at 265, 325, 385, 466, 592, and 640 nm, respectively. The initial three bands were within the ligand while the other bands were resulting from charge transfer and 6A1→4T1 transitions indicating distorted octahedral geometry around the iron(III) metal ion (Mangamamba et al., 2014). Chromium(III) complex (9) showed six bands at 285, 305, 380, 465, 562, and 634 nm, respectively. The first three bands were appointed to 4A2g→4T1g(F), 4A2g→4T2g, and 4A2g→2T2g transitions, respectively, demonstrating octahedral structure around the Cr(III) ion. Iron(III) complex (8) and chromium(III) complex (9) had magnetic moment values of 5.2 and 3.8B.M. suggestive of a high spin octahedral structure (Mangamamba et al., 2014). Zn(II)-based complex (11) gave bands of octahedral structures and diamagnetic which gave merely intraligand transitions (supporting information table 4).
3.6 Thermal studies [DTA and TGA]
As the IR spectral data indicated the existence of water molecules, thermal analyses (DTA and TGA) were done to assure their character which was listed in supporting information table 5. The thermal curves in 27–800 °C temperature range for complexes (2–11) revealed that these complexes were thermally stable up to 35 °C. Dehydration was illustrated by endothermic peaks within 70–90 °C temperature range, corresponding to loss of water molecules (Surati, 2011; El-Tabl et al., 2016). The decomposition mechanism of ligand 1 was illustrated in Fig. 3.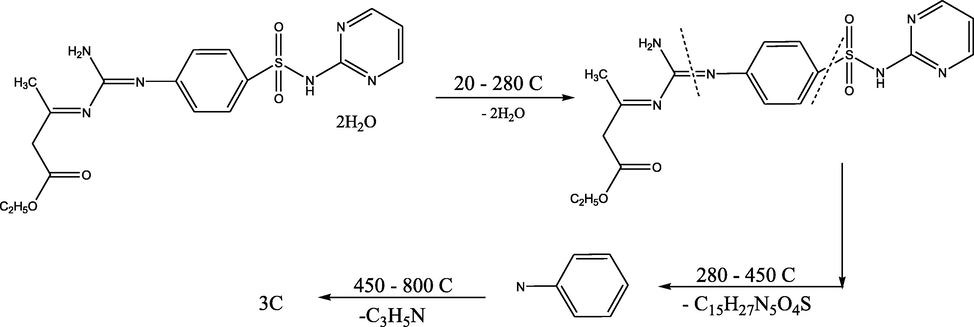
Thermal decomposition mechanism of the ligand 1.
The decomposition step for complex (2) started with the break of H-bonding associated with an endothermic peak at 45 °C, followed by the loss of two coordinated water molecules with an endothermic peak at 120 °C and with 8.96% weight loss (Calc. 9.01%). A peak was observed at 170 °C with 20.84% weight loss (Calc. 20.78%). It was owing to two acetate groups. The endothermic peak found at 355 °C might be induced by the melting point. Oxidative thermal decomposition occurred in 465–650 °C temperature range with exothermic peaks leaving CuO 17.33% weight loss (Calc. 17.77%) (Kumar et al., 2014; El-Tabl et al., 2016). The decomposition step for complex (3) began with the disruption of H-bonding with endothermic peak at 45 °C, followed by the elimination of two water molecules at 83 °C with weight loss of about 8.65% (Calc. 5.57%), and then by the removal of two coordinated water molecules with a peak at 135 °C and 9.03% weight loss (Calc. 9.04%). Another thermal decomposition peaked at 215 °C with weight loss of approximately 17.55% (Calc. 17.66%) as a result of a coordinated sulphate group loss. An additional endothermic peak was noticed at 320 °C with no weight loss which could be caused by the melting point. Finally, the complex showed exothermic peaks in the 440–660 °C temperature range corresponding to oxidative thermal decomposition which proceeded gradually with a final residue at 660 °C which was allocated to CuO (Kumar et al., 2014; El-Tabl et al., 2016) (see Figs. 4 and 5).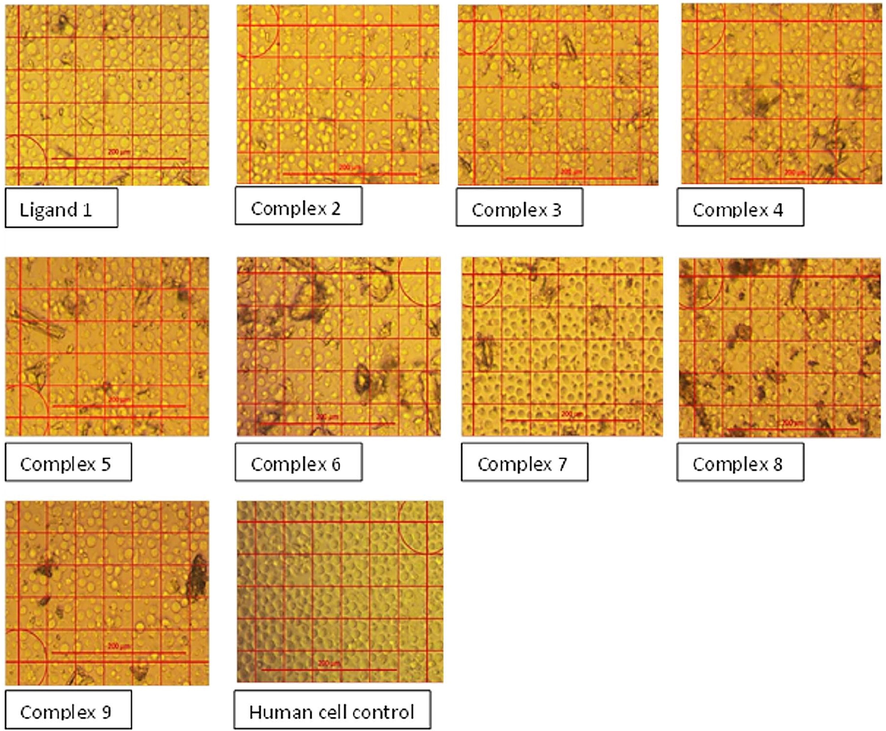
Inverted microscopic examination of the human mononuclear cells from peripheral blood treated with a concentration of 62.5 µg/mL of the ligand and the complex compounds.
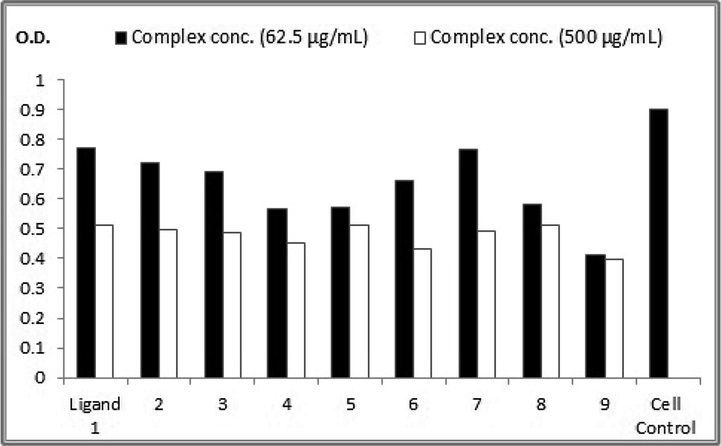
Crystal violet stain colorimetric assay at OD = 590 nm for the nine complexes in compare to control human hepatoma (HepG2) cell line.
The decomposition step for complex (4) was initiated with H-bonding break associated with an endothermic peak identified at 48 °C, followed by the loss of a hydrated water molecule (H2O) at 85 °C associated with 3.08% weight loss (Calc. 3.04%) and then by the deletion of two coordinated water molecules with a peak at 145 °C and 6.46% weight loss (Calc. 6.27%). Yet, the endothermic peak observed at 177 °C accompanied by 13.27% weight loss (Calc. 13.18%) was due to two chloride groups. A peak was detected at 345 °C resulting from the melting temperature point. Oxidative thermal decomposition occurred in 455–670 °C temperature range with exothermic peaks, leaving CuO with about 17.02% weight loss (Calc. 17.00%) (Surati, 2011; El-Tabl et al., 2016). The decomposition step for complex (5) began with the break of H-bonding with a peak at 45 °C, followed by the loss of 2 H2O at 87 °C with weight loss of about 5.75% (Calc. 5.69%). Then, two coordinated water molecules were removed with an endothermic peak at 135 °C and 6.06% weight loss (Calc. 6.04%). A peak spotted at 165 °C, with weight loss of about 22.18% (Calc. 22.14%), was designated to two nitrate groups. The peak at 310 °C might be generated by the melting point. Oxidative thermal decomposition occurred in 440–680 °C temperature range with exothermic peaks, leaving CuO with about 18.42% weight loss (Calc. 18.32%) (Kumar et al., 2014; El-Tabl et al., 2016).
The decomposition step for complex (6) started with H-bonding break which was associated with an endothermic peak at 46 °C. This was followed by the removal of 2H2O molecules at 87 °C with weight loss of about 5.57% (Calc. 5.51%). Then, two coordinated water molecules were eliminated with a peak at 137 °C and 5.86% weight loss (Calc. 5.84%). The peak which was observed at 186 °C with 20.35% weight loss (Calc. 20.30%) was attributed to two acetate groups. An endothermic peak was detected at 326 °C because of the melting point. Oxidative thermal decomposition occurred in the 460–680 °C temperature range with exothermic peaks, leaving CoO weight loss of about 16.13% (Calc. 16.18%) (Kumar et al., 2014; El-Tabl et al., 2016).
The decomposition step for complex (7) was initiated with H-bonding disruption with a peak at 46 °C, followed by the rejection of three H2O at 89 °C with about 8.86% weight loss (Calc. 8.73%). Then, two coordinated water molecules were lost through a peak at 125 °C with weight loss of about 6.34% (Calc. 3.38%). Nevertheless, the peak observed at 180 °C associated with 22.03% weight loss (Calc. 22.32%) was caused by two acetate groups. The peak at 350 °C was possibly a melting point. Oxidative thermal decomposition occurred in the 465–660 °C temperature range with exothermic peaks, leaving NiO with weight loss of about 18.23% (Calc. 18.17%) (Kumar et al., 2014; El-Tabl et al., 2016).
The decomposition step for complex (8) was triggered with the break of H-bonding with a peak at 47 °C which was followed by the removal of 2H2O at 86 °C with 5.77% weight loss (Calc. 5.57%). Then, three coordinated water molecules were eradicated with a peak at 120 °C and 8.55% weight loss (Calc. 8.86%). A peak at 185 °C associated with 17.03% weight loss (Calc. 17.27%) was owing to sulphate group. The endothermic peak at 359 °C was a melting temperature point. Oxidative thermal decomposition occurred in the 460–650 °C temperature range associated with exothermic peaks, leaving Fe2O3.
The decomposition step for complex (9) began with H-bonding split accompanied by an endothermic peak at 45 °C with the elimination of 3H2O at 84 °C and with 8.06% weight loss (Calc. 8.18%). Three coordinated water molecules were eliminated through a peak at 130 °C with weight loss of about 8.76% (Calc. 8.91%). The peak noticed at 180 °C with 17.22% weight loss (Calc. 17.39%) was attributed to one sulphate group. The peak at 350 °C was caused by the melting point. Oxidative thermal decomposition occurred in the 450–670 °C temperature range concomitant with exothermic peaks leaving Cr2O3.
The decomposition step for complex (10) was initiated with H-bonding disruption with a peak at 44 °C. Then, 2H2O were lost at 87 °C with weight loss of about5.34% (Calc. 5.55%). Afterwards, three coordinated water molecules were removed through a peak at 129 °C with 8.45% weight loss (Calc. 8.81%). On the other hand, the peak spotted at 184 °C with 17.23% weight loss (Calc. 17.24%) was resulting from one sulphate group. The endothermic peak appeared at 352 °C was a melting temperature point. Oxidative thermal decomposition occurred in the 440–660 °C temperature range accompanied by exothermic peaks leaving NiO.
The decomposition step for complex (11) started with H-bonding collapse with an endothermic peak at 48 °C before the loss one H2O at 80 °C with 2.87% weight loss (Calc. 2.73%). Later, two coordinated water molecules were removed at 137 °C with 5.46% weight loss (Calc. 5.61%). A peak detected at 175 °C with weight loss of about 19.23% (Calc. 19.49%) was caused by two acetate groups. The melting point triggered a peak at 335 °C. Oxidative thermal decomposition occurred in the 440–660 °C temperature range with exothermic peaks leaving NiO 16.56% weight loss (Calc. 16.69%).
3.7 Electron spin resonance (ESR)
The ESR spectral data for metal complexes (2), (4), and (9) are stated in (supporting information table 6). The spectra of Cu(II) metal complexes (2) and (4) were representative of species d9 conformation having an axial type of a d(x2-y2) ground state which is the most frequent type for Cu(II) complexes (Akbar et al., 2011). The metal-based compounds had g|| > g⊥ > 2.0023 demonstrating an octahedral geometry around the Cu(II) metal ion (El-Tabl et al., 2011). G-values = (g||-2)/(g⊥-2). If G is>4.0, local tetrahedral axes are misaligned parallel or only slightly misaligned and if G is below 4.0, this means that there is a significant exchange coupling (Thomas and Mohanan, 2017). The g||/A|| values of the new complexes lied within the range expected for the square planar or octahedral metal complexes. The orbital reduction factors (K||, K⊥, K), which are measures of covalency, were also calculated. Concerning the Cu(II) complexes (2) and (4), the K values signified a covalent bond character (El-Razek et al., 2020). Also, the g-values confirmed the covalent bond character. The in-plane σ- covalency parameter, α2 (Cu) was suggestive of a covalent bonding. The complexes (2) and (4) displayed β1 values i.e. a covalence character in the in-plane π- bonding. While β2 for the complexes denoted an ionic bonding character in the out of plane π- bonding. However, complex (4) had an ionic bond character out of the plane bonding (Thomas and Mohanan, 2017; El-Razek et al., 2020). The Cu(II) complexes calculated orbital populations (a2d) implied a d(x2-y2) ground state. Cr(III) complex (9) showed isotropic spectra with 2.09 value.
3.8 Chemotherapeutic effect
To assess the cytotoxicity and the anticancer effectiveness of the new complexes, the IC50 values (the concentration that decreases the cell viability to 50% of the cell control viability) were determined. These values were calculated utilizing human mononuclear cells incubated with the concentrations of 1000, 500, 250, 125, 62.5, and 31.2 µg/mL of the ligand and compounds for 36 h and visually, the viable cells were counted under the inverted microscope (Fig. 2).
The complexes 1, 3, and 5 showed rare cytotoxic activities while complexes 2, 4, 6, 7, 8, and 9 were weakly cytotoxic (Table 7) (see Tables 8 and 9).
Complex No.
Clear zone (ϴmm)
Pseudomonas aeuginosa
Staphylococcus aureus
Candida albicans
A. niger
Neomycin
28
24
18
0
Cycloheximide
0
0
0
34
(1)
14
14
21
25
(2)
22
18
24
21
(3)
19
30
25
28
(4)
19
25
14
16
(5)
18
15
21
15
(6)
20
20
24
29
(7)
26
23
26
30
(8)
19
25
24
19
(9)
15
28
19
20
##
Complex conc. (62.5 µg/mL)
Viability percentage at conc. (62.5 µg/mL)
Complex conc. (500 µg/mL)
Viability percentage at conc. (500 µg/mL)
Ligand 1
0.771
85.28
0.511
56.52
2
0.72
79.64
0.499
55.19
3
0.69
76.32
0.486
53.76
4
0.566
62.61
0.45
49.77
5
0.571
63.16
0.513
56.74
6
0.66
73.00
0.433
47.89
7
0.766
84.73
0.49
54.20
8
0.58
64.15
0.513
56.74
9
0.411
45.46
0.397
43.91
Cell Control
0.904
In vitro growth inhibition assays were done on human cancer cell line (HepG2) at various concentrations in comparison to the non-treated HepG2 control. A highly significant anticancer effect, against HepG2 cells, after 24 h exposure, was specified by the crystal violet stain colorimetric assay. The anticancer effects of the ligand and the new metal-containing compounds were directly proportional to their concentration. The Cr(III)-, Fe(III)-, and Cu(II)-based complexes, in order, had the highest anticancer activities while Ni(II) had the lowest activity in comparison to the ligand (Fig. 3). These results were in agreement with Alturiqi et al. (2018) who found that Cr(III) complex exhibited higher in vitro cytotoxicity against both the selected cancer and normal cell lines when compared to the ligand while the Ni(II) and Cu(II) complexes displayed lower anticancer activities than that of the ligand.
3.9 Antimicrobial activity
The antimicrobial activities of the ligand (1) and of its metal complexes were evaluated in vitro by investigating their antibacterial and antifungal effects. The results were compared with Tetracycline which is the standard medication. S. aureus, P. aeruginosa, C. albicans, and A. niger were utilized as bacterial, yeast, and fungal test organisms. The ligand and its metal complexes (2–9) displayed antibacterial and antifungal effects against these life forms (supporting information Table 7). It was detected that the metal-based complexes had a more noteworthy effect than the ligand on the test organisms. Ligand (1) showed direct antibacterial and antifungal activities in comparison to the standard antibacterial (Neomycin) and antifungal (cycloheximide) drugs with clear zones values of 14, 14, 16, and 16 mm for S. aureus, p. aeruginosa, C. albicans and A. niger, respectively. Complexes (2), (5), (4), (3), and (7) were more hostile to G +ve bacteria than ligand (1) with clear zone values of 22, 18, 19, and 19 mm, respectively. Besides, complexes (2), (3), (6), (8) and (9) had higher antibacterial effects on G-ve bacteria than the ligand with clear zone values of 18, 15, 25, 30, 20, and 23 mm, respectively. Moreover, complexes (2), (4), (3), (6), (7), (8), and (9) revealed antifungal activities against the yeast C. albicans which were more elevated than the ligand with values 21, 24, 21, 19, 25, 24, and 26 mm, respectively. Concerning the antifungal properties of both ligand and complexes against A. niger, complexes (6) and (7) exceeded the ligand antifungal activity with clear zone values of 29 and 30 mm, respectively.
4 Conclusion
New sulfadiazine Schiff base ligand-based metal complexes were successfully synthesized. The data obtained from the analytical and physicochemical analyses confirmed the stability of the new compounds. The ESR spectral data of solid Cu (II) complexes exposed an axial type of a d(X2-y2) ground state with a covalent bond nature. A distorted octahedral geometry was adopted by the new complexes around the metal ion. The ligand and its complexes showed augmented cytotoxic activity, against human liver cancer cells, compared to the standard drug; Vinblastine sulfate. The metal complexes showed a higher chemotherapeutic activity against cancer cells than the free ligand indicating enhanced antitumor activity upon coordination. This could be a step forward to apply these complexes clinically on experimental models as therapeutic agents for liver cancer.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Transition metal complexes of a multidentate Schiff base ligand containing guanidine moiety: synthesis, characterization, anti-cancer effect, and anti-microbial activity. J. Mol. Struct.. 2020;1203:127381
- [Google Scholar]
- Synthesis, characterization and antibacterial studies of metal complexes of sulfadiazine with N-alkyl-N-phenyldithiocarbamate. Bull. Chem. Soc. Ethiop.. 2013;27:77-84.
- [Google Scholar]
- Mixed-ligand ternary complexes of potentially pentadentate but functionally tridentate Schiff base chelates. Polyhedron. 2011;30:542-548.
- [Google Scholar]
- Alturiqi, A.S., Alaghaz, A.N.M.A., Ammar, R.A., Zayed, M.E, 2018. Synthesis, spectral characterization, and thermal and cytotoxicity studies of Cr(III), Ru(III), Mn(II), Co(II), Ni(II), Cu(II), and Zn(II) complexes of schiff base derived from 5-hydroxymethylfuran-2-carbaldehyde, J. Chem.
- Synthesis, characterization and antimicrobial activity of salicylaldehyde benzenesulfonylhydrazone (Hsalbsmh) and its Nickel(II), Palladium(II), Platinum(II), Copper(II), Cobalt(II) complexes. Inorg. Chem. Comm.. 2011;14:1550-1553.
- [Google Scholar]
- Proton chemical shifts in NMR spectra of diamagnetic metal-aminopolycarboxylate complexes. Inorg. Chim. Acta. 1980;9:233-235.
- [Google Scholar]
- Metal-based sulfonamides: synthesis, characterization, antibacterial, antifungal and cytotoxic properties of pyrrolyl- and thienyl-derived compounds. Appli. Organomet. Chem.. 2007;21:728-738.
- [Google Scholar]
- Transition metal complexes of a multidentate Schiff base ligand containing guanidine moiety: synthesis, characterization, anti-cancer effect, and anti-microbial activity. J. Mol. Struct.. 2020;1203:127381
- [Google Scholar]
- Spectroscopic characterization and biological activity of metal complexes with an ONO trifunctionalized hydrazone ligand. J. Coord. Chem.. 2008;61:2380-2401.
- [Google Scholar]
- Synthesis, characterization and esr studies of new copper(ii) complexes of vicinal oxime ligands. J. Korean Chem. Soc.. 2011;55:603-611.
- [Google Scholar]
- Novel metal complexes of guanidine ligand; synthesis, spectroscopic characterization and biological activity. Int. J. Chem. Tech. Res.. 2013;5:430-449.
- [Google Scholar]
- Synthesis, structural characterization and cytotoxic activity of novel symmetrical and asymmetrical metal complexes of guanidinium Schiff base. J. Chem. Biol. Phys. Sci. Section A. 2015;5:3875-3908.
- [Google Scholar]
- Antibacterial evaluation of ethoxy-oxime schiff base ligand and its metal complexes. Asian J. Sci. Technol.. 2016;7:3167-3180.
- [Google Scholar]
- Synthesis and characterization of a hydrazone ligand containing antipyrine and its transition metal complexes. J. Coord. Chem.. 2008;61:1983-1996.
- [Google Scholar]
- Synthesis, spectral characterisation, electrochemical, and fluorescence studies of biologically active novel Schiff base complexes derived from E-4-(2-hydroxy-3-methoxybenzlideneamino)-N-(pyrimidin-2-yl)benzenesulfonamide. Turk. J. Chem.. 2014;38:521-530.
- [Google Scholar]
- A novel approach of potent antioxidant and antimicrobial agents containing coumarin moiety accompanied with cytotoxicity studies on the newly synthesized derivatives. J. Appl. Pharm. Sci.. 2017;7:186-196.
- [Google Scholar]
- Synthesis, characterization, and biotoxicity of N N donor sulphonamide imine silicon(IV) complexes. Bioinorg. Chem. Appl.. 2006;2006:13743.
- [Google Scholar]
- O-H hydrogen bonding promotes H-atom transfer from α C-H bonds for C-alkylation of alcohols. Science. 2015;349:1532-1536.
- [Google Scholar]
- Synthesis, characterization and thermogravimetric analysis of Co(II), Ni(II), Cu(II) and Zn(II) complexes supported by ONNO tetradentate Schiff base ligand derived from hydrazino benzoxazine. J. Saudi Chem. Soc.. 2017;21:S457-S466.
- [Google Scholar]
- Syntheses and structural studies on coordination compounds of polystyrene-anchored schiff base with some metal ions. J. Chil. Chem. Soc.. 2014;59:2260-2264.
- [Google Scholar]
- Lewis, J.E., Wilkins, R.G., 1967. Modern Coordination Chemistry: Principles and Methods. Interscience Publishers, New York.
- Characterization of complexes synthesized using Schiff base ligands and their screening for toxicity two fungal and one bacterial species on rice pathogens. Int. J. Inorg. Chem.. 2014;2014:1-22.
- [Google Scholar]
- Oxovanadium(IV) complexes of bioinorganic and medicinal relevance: Synthesis, characterization and 3D molecular modeling of some oxovanadium(IV) complexes involving O, N-donor environment of salicylaldehyde-based sulfa drug Schiff bases. Arab J. Chem.. 2016;9:S1084-S1100.
- [Google Scholar]
- Studies on some transition metal complexes of Schiff bases derived from quinoxaline-2-carboxaldehyde. Synth. React. Inorg. Met. Org. Chem.. 2003;33:81-496.
- [Google Scholar]
- Biological studies and characterisation of sulfa drug based ligand and transition metal complex. J. Chem. & Cheml. Sci.. 2017;7:947-951.
- [Google Scholar]
- Synthesis, characterization and biological activity of some transition metals with Schiff base derived from 2-thiophene carboxaldehyde and aminobenzoic acid. Spectrochim. Acta A Mol. Biomol. Spectrosc.. 2005;62:1140-1150.
- [Google Scholar]
- Metal complexes in cancer therapy - an update from drug design perspective. Drug Des. Devel. Ther.. 2017;11:599-616.
- [Google Scholar]
- Elangovana, Synthesis, characterization and biological studies of sulfadiazine drug based transition metal complexes. J. Chem. Pharm. Res.. 2016;8(8):202-211.
- [Google Scholar]
- The comparison of MTT and CVS assays for the assessment of anticancer agent interactions. PLoS ONE. 2016;11:155772
- [Google Scholar]
- Resveratrol ameliorates oxidative stress and organ dysfunction in Schistosoma mansoni infected mice. Exp. Parasitol.. 2017;174:52-58.
- [Google Scholar]
- Synthesis, spectroscopy and biological investigations of manganese(III) Schiff base complexes derived from heterocyclic b-diketone with various primary amine and 2,2'-bipyridyl. Spectrochim. Acta Part A Mol. Biomol. Spectrosc.. 2011;79:272-277.
- [Google Scholar]
- Design, synthesis, and docking of sulfadiazine schiff base scaffold for their potential claim as inha enoyl-(Acyl-carrier-protein) reductase inhibitors. Asian J. Pharm. Clin. Res.. 2018;11:233-237.
- [Google Scholar]
- Synthesis and biological evaluation of some 3d metal complexes with a novel heterocyclic Schiff base. J. Chinese Chem. Soc.. 2017;64:1510-1523.
- [Google Scholar]
- Screening methods to determine antibacterial activity of natural products. Braz. J. Microbiol.. 2007;38:369-380.
- [Google Scholar]
- Vogel A.I., 1966. A text-book of quantitative inorganic analysis, including elementary instrumental analysis, by Arthur I. Vogel, [London] Longmans.
- Comparative analysis of desalination technologies. Energy Procedia. 2014;61:2604-2607.
- [Google Scholar]
- Synthesis and characterization of new Schiff base metal complexes and their use as catalysts for olefin cyclopropanation. Phosphorus Sulfur Silicon Relat. Elem.. 2010;185:785-798.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2020.08.010.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary Data 1
Supplementary Data 1







