Non-specific adsorption of serum and cell lysate on 3D biosensor platforms: A comparative study based on SPRi
⁎Corresponding authors at: Center for Drug Delivery System, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, People’s Republic of China (V. Singh). National Center for Nanoscience and Technology, Beijing 100190, People’s Republic of China (J. Zhu). Tel./fax: +86 21 20231980. kasana.chem@gmail.com (Vikramjeet Singh), jizhu@nanoctr.cn (Jingsong Zhu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Comparative study for non-specific adsorption of cell lysate and serum on recently developed biosensor surfaces is reported. Different surface chemistries including, polyethylene glycol (PEG), α-cyclodextrin (CD), hydrogel dextran and surface initiated polymerization (SIP) based gold surfaces were taken into account for this study. Various techniques including, surface plasmon resonance imaging (SPRi) and matrix-assisted laser desorption ionization tandem time-of-flight mass spectrometry (MALDI−TOF/TOF MS) technique to evaluate the surfaces with Fourier transform infrared spectroscopy (FTIR) for the confirmation of surface fabrication were used. A high non-specific adsorption response of cell lysate and serum was observed on these so-called non-fouling surfaces. The obtained results from this comparative study provided some hope to explore SIP and dextran surface as a universal platform for biosensor applications. SIP produced best result and showed high sensitivity and minimum non-specific adsorption. We believe that this comparative study will surely help the researchers to further upgrade these surfaces and especially SIP to make it more universal for biosensor microarray applications including biomarker discovery in high throughput format.
Keywords
Non-specific adsorption
Cell lysate and human serum
α-cyclodextrin
Dextran
Surface initiated polymerization
Surface plasmon resonance imaging
1 Introduction
SPR imaging technology has proven to be an invaluable tool for high-throughput analysis of bio-molecular interactions (Kodoyianni et al., 2011) such as, drug discovery (Singh et al., 2014,2019), biomarker screening (Shabani et al., 2013), nucleic acid detection (Roberta et al., 2013), food safety analysis (Piliarik et al., 2009), environmental analysis (Mauriz et al., 2007), DNA hybridization (Ananthanawat et al., 2010 and Malic et al., 2009), protein–DNA interaction (Pillet et al., 2013) and living cell activation (Hiragun et al., 2012). SPR sensing has been significantly developed in recent years to quantify biomarkers in complex matrixes, such as serum (Choi et al., 2010; Su et al., 2008 and Ladd et al., 2009), plasma (Teramura et al., 2007), saliva (Yang et al., 2005) and cell lysate (Kyo et al., 2005). SPR biosensors have been shown to be suited for the detection of several cancer biomarkers (Sankiewicz et al., 2015) in solubilized samples, such as oropharyngeal squamous cell carcinoma (Liu et al., 2012), prostate cancer (Choi et al., 2010), colorectal, gastric, and pancreatic cancer (Su et al., 2008), intestinal cancer (Ladd et al., 2009), liver cancer (Teramura et al., 2007) and ovarian cancer (Liu et al., 2012) among others. However, serum and cell lysate screening on SPRi provide real time detection results in less time with low human error, but due to their protein and lipid rich matrix respectively, they expected to significantly foul on sensor surface.
Hence, non-specific adsorption is a major roadblock to analyze complex biological samples such as cell lysates by using label-free biosensor technologies (Vaisocherova et al., 2008). The most promising surface chemistries include polycarboxybetaine acrylamide surface (Brault et al., 2012) and polyMeOEGMA (Rodriguez et al., 2011) surfaces. Recently, a peptidoid polymer (Lin et al., 2011) and a peptide (Bolduc et al., 2011) self-assembled monolayer (SAM) were proposed to reduce nonspecific adsorption of serum. All of these surfaces have been developed to prevent non-specific adsorption of proteins from serum, but to our knowledge, little information is available about low-fouling surface chemistry for cell lysate. As a result of the lipid-rich nature of cell lysate in opposition to the protein rich composition of serum, it is expected that the surfaces developed to prevent non-specific adsorption from serum will not be effective when used with cell lysate (Aube et al., 2013). A recent review details the significant efforts made to minimize the non-specific adsorption problem in biosensing techniques (Reimhult et al., 2015). Starts from the 2D polyethylene glycol (PEG) surface, a number of 3D matrixes have been proposed by researchers for biosensing applications based on hydrogel dextran (LofAs et al., 1990), surface initiated polymerization (Ma et al., 2010) and α-cyclodextrin (He et al., 2013) on gold substrate. Researchers claimed the high loading capacity and enhanced sensitivity and non-fouling nature of all these surfaces without providing much data related to non-specific adsorption study. Since non-specific adsorption is a key problem and forbids the broad application spectrum of biosensors, it is necessary to find an efficient approach to solve this issue. Apart from the type of biosensors, substrates and analytes used, surface chemistry plays the most important role in non-specific adsorption. In light of surface chemistry importance, above described various types of surfaces were fabricated and characterized by infrared spectroscopy. After, all surfaces were evaluated for non-specific adsorption of stem cell lysate (induced pluripotent stem cell) and human serum before and after carboxylation of SAM by using surface plasmon resonance imaging (Fig. 1). Furthermore, non-specifically bound lipids/proteins were identified by using matrix-assisted laser desorption ionization (MALDI) time-of-flight (TOF) MS onto the biosensor surfaces.
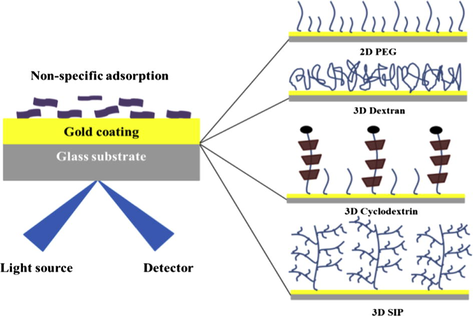
- Schematic for NSA comparative study on 4 different surfaces.
2 Experimental section
2.1 Reagents
Unless otherwise noted, material and solvents were obtained from commercial suppliers and used without further purification: Gold coated slides (Plexera), SH-(PEG)n-OH, (M.W. 2000) and SH-(PEG)n-OCH3 (M.W. 1000) (Shanghai Yan Yi biotech. China); and α-cyclodextrin (M.W. 1000), EDC-HCl (1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride), NHS (N-hydroxy succinimide) and 2-(2-aminoethoxy)ethanol (EG2-NH2) (Aladdin Chemistry). Succinic anhydride, 2,2 ethylenedioxy-bis (ethylamine) and 4-(dimethylamino) pyridine (DMAP) were purchased from Aldrich. ω-Mercaptoundecyl bromoisobutyrate was purchased from HRbio (Beijing). 2,2-Bipyridyl (Bipy), Copper(II) chloride (CuCl2), 2-Hydroxyethyl methacrylate (HEMA), poly(2-hydroxyethyl methacrylate) (OEGMA), Ascorbic acid (AscA), N,N-Disuccinimidyl carbonate (DSC), 4-(Dimethylamino) pyridine (DMAP), epichlorohydrin, and bovine serum albumin (BSA) were purchased from sigma Aldrich. Dextran (T-500) was purchased from Pharmacia (Beijing).
2.2 Preparation of 2D PEG surface
The 2D PEG surface was fabricated as per our previous published work (Singh et al., 2014,2019). In Brief, PlexArray® gold coated chips were cleaned with a plasma cleaner and ethanol. The chips were then kept overnight in ethanolic solution 1 mM (1:10 ratio) of SH-(PEG)n-OH (M.W. 2000) and SH-(PEG)n-OCH3 (M.W. 1000) to prepare dense SAM of PEG. The slides were washed with ethanol for 30 min under vigorous shaking to remove the unbound particles. The slides were treated with DMF solution of succinic anhydride and DMAP for 16 h at room temperature to obtain carboxy terminated PEG surface.
2.3 Preparation of 3D SIP surface
A mixed SAM solution was prepared by initiators ω-mercaptoundecyl bromoisobutyrate (BrC(CH3)2COO(CH2)11SH) and EG3-thoil in 1:99 ratio. The chips were immersed in this mix (1 mM total concentration) for 16 h at room temperature, and then thoroughly washed by ethanol and Milli-Q water and dried in a nitrogen stream. Polymerization solution was prepared by 64 mg Bipy, 10 ml 0.04 M CuCl2, 2.6 g HEMA, 7.2 g OEGMA, 20 ml Milli-Q water and 20 ml methanol. After 30 min deoxygenation, 10 ml of AscA (0.04 M) was added to the solution and the chips were immersed in this solution for 16 h at room temperature under an atmosphere of argon. After being thoroughly washed with methanol and Milli-Q water, the chips were incubated in a DMF solution containing 0.1 M DSC and 0.1 M DMAP for 16 h for acidification.
2.4 Preparation of cyclodextrin surface
The 2D PEG surface was fabricated as per above explained procedure. For the preparation of 3D cyclodextrin matrix, PEG assembled slides were treated with aqueous α-cyclodextrin solution for two hours at room temperature. After threading the cyclodextrins to PEG chain, the terminal carboxyl group was activated by EDC (0.39 M) and NHS (0.2 M) mixture followed by capping of chain by Z-Tyr-OH group. Finally, the cyclodextrins hydroxy groups were converted into carboxyl by DMF solution of succinic anhydride and DMAP for 16 h at room temperature under shaking (Singh et al., 2014,2019).
2.5 Preparation of dextran hydrogel surface
Dextran hydrogel surface was fabricated by previously published work (LofAs et al., 1990). Gold chips after cleaning were self-assembled with hydroxy-terminated polyethylene glycol (EG3) in ethanol at 4 °C for overnight. The resulting hydrophilic surface was then reacted with a 0.6 mol dm−3 solution of epichlorohydrin in a 1:1 mixture of 0.4 mol dm−3 sodium hydroxide and diglyme for 4 h at 25 °C. After thoroughly washing with water, ethanol and water again, the surface was treated with a basic dextran solution [3.0 g dextran T500 (Pharmacia AB) in 10 cm3 of 0.1 mol dm−3 sodium hydroxide] for 20 h at 25 °C. And finally, hydroxy groups of dextran surface were converted into carboxyl one by using same solution of DMF and DMAP as used for SIP and CD.
2.6 FTIR spectroscopy method
To prove the fabrication of each surface, we employed a Fourier transform infrared (FTIR) spectrophotometer (GX-Perkin Elmer, USA) using the reflection mode at a resolution of 4 cm−1 over the 4000–400 cm−1 spectral region to reveal the chemical bond and functional groups such as carbonyl (C⚌O) and hydroxyl (OH) groups belonging to COOH-terminated alkanethiol SAMs. FTIR spectrums of all above prepared, PEG, 3D CD, 3D SIP and 3D dextran films modified on gold were recorded on FTIR spectrometer (Perkin Elmer) with 64 scans at a resolution of 4 cm−1.
2.7 Cell lysate preparation
Stem cell lysate was obtained by using the same method as our previous published work (Nand et al., 2014). R1 mouse embryonic stem cells (mESCs) were homogenized with lysate buffer. Embryonic cell lysate was prepared in two lysate buffer separately: Tris-HCl buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% SDS, pH 7.4) and HEPES buffer (20 mM HEPES, 150 mM NaCl, 0.05% Tween 20, pH 7.4) containing protease and phosphatase inhibitor cocktail (Roche, 04693124001). The cell lysates were centrifuged at 12,000 rpm for 20 min at 4 °C, and the supernatants were collected and used for SPRi analysis. Total protein content was determined by Bradford protein assay. Cell lysates were diluted to reduce the bulk effect.
2.8 SPRi analysis
All the experiments were carried out using the PlexArray® HT system (Plexera, LLC) which is based on surface plasmon resonance imaging. Oval regions of interest (ROIs) were set as 25 pixels × 20 pixels area in imaging area. Serum and cell lysate (1:100 dilution) containing Tween 20 (0.05%), pH 7.4 were used as analytes with an association and dissociation flow rate of 3 μl/s (25 °C) at different concentrations by serial dilution. HEPES (10 mM Hepes, 150 mM NaCl, 1 mM MgCl2, and 1 mM CaCl2, pH 7.4) containing Tween 20 (0.05%) was used as a running buffer. A solution of NaOH (20 mM) was used to regenerate the surface (300 s) and remove bound cell lysate from the lectin microarray, enabling the sensor chip to be reused for additional analyte injections. The 10 ROIs were selected at different places to cover the whole surface and ensure the analysis uniformity. Average sensitivity data from all 10 spots were provided with standard deviation. Data were analyzed according to our previous work (Singh et al., 2014,2019).
2.9 MALDI–TOF analysis of adsorbed proteins
MALDI−TOF/TOF analyses were performed to monitor lipids/proteins on the different surfaces using α-cyano-4-hydroxycinnamic acid (CHCA, with peptides and lipids in positive mode) and 2,6-dihydroxyacetophenone (DHA, with lipids in negative mode) matrixes. Matrix solutions were prepared at 10 mg/mL in 1:1 acetonitrile H2O + 0.1% trifluoroacetic acid. It is noteworthy to point out that the slides used for MS experiments were 25 × 75 × 1.0 mm microscope slides to fit the slide adaptor of the MS instrument. A 30 min trypsin digestion was run at 37 °C on three different spots (3 μL) per slide on a heating plate controlled by a temperature sensor. The substrate was mounted on hydrated cotton wool on the heating plate to distribute heat uniformly. A reference gold-coated slide with a drop of water was used as a reference for the temperature sensor. Matrix deposition was performed by successive evaporation of 0.5 μL of matrix solution on the digested spots to analyze proteins and beside the digested spots for lipid analysis. MS analysis of the adsorbed material was performed with a MALDI−TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) as previously described (Ratel et al., 2011).
3 Results
3.1 Surface characterization by FTIR spectroscopy
Prior to use for SPRi experiments, all fabricated chips (after carboxylation) were characterized by FTIR for structure confirmation (Fig. 2). In the IR spectrum, we observed characteristic peaks at 1160 cm−1 which correspond to C—O—C stretches in the PEG chain for all 4 surfaces. The increase in intensity of the peak at 1160 cm−1 was observed, after the PEG SAM was further modified in order to fabricate other three surfaces, α-CD, dextran and SIP. The new peak at 1735 cm−1 (more intense in case of SIP and dextran) shown corresponds to the carboxyl group C⚌O vibration. Presented IR data were recorded after carboxylation showing complete missing of hydroxyl O—H peak at 3200–3500 cm−1 region and showing carboxyl O—H stretching at 2900 cm−1. In whole, FTIR data suggested the successful fabrication of all four surfaces.
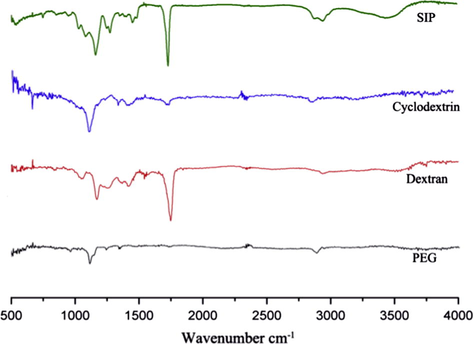
- The FTIR spectra of all 4 surfaces include, polyethylene glycol, α-cyclodextrin, dextran and SIP.
3.2 SPRi analysis
All 4 types of well characterized gold slides mounted on SPRi instrument and whole process were carried out on the same, starting from activation by EDC/NHS to flow of serum and cell lysate through flow cell. As shown in Fig. 3A, after activation with EDC/NHS mixture (0.39:0.1 M) for 15 min, surfaces were thoroughly blocked by EG2-NH2 (1 M) and then BSA (100 μg/ml) 10 min each followed by successive washing of 5 min by running buffer (20 mM HEPES). After blocking step, printing area was exposed to serum and cell lysate (separated for 200 s by single regeneration step (20 mM NaOH)). Immobilization amounts of NHS were measured (Fig. 3B) for all slides and suggested the high loading capacity of 3D SIP and 3D dextran than 3D CD which own higher than 2D PEG as expected. Our conclusion is in agreement with the previously reported (Singh et al., 2014,2019) studies that 3D has high immobilization capacity and owns more functional groups which is confirmed and compared in this study.
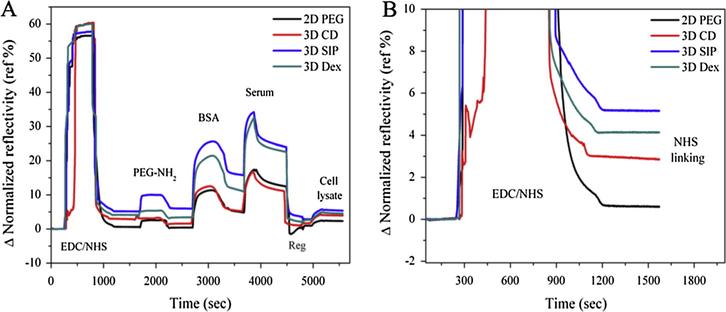
- A complete experiment setup (A) showing EDC/NHS activation of carboxyl groups followed by blocking by PEG-NH2 and bovine serum albumin (BSA) and then flow of embryonic stem cell lysate and human serum in order to check the non-specific adsorption on all 4 surfaces (B) a sensorgram revealing NHS immobilization capacity of each surface after activation with EDC/NHS.
3.3 SPRi analysis of adsorption of serum and cell lysate before carboxylation
As it is well known, that hydroxy functionalized biosensor slides own less non-specific adsorption of serum and lysate, we compared these 4 surfaces before carboxylation step using SPRi. Here, slides were blocked by BSA only for 10 min on instrument and then flow serum and miPSC lysate as described above. Adsorption behaviors of serum and cell lysate are shown in Fig. 4A and B respectively. We note that, the association behavior of serum is approximately same when compared with each slide including 2D PEG, 3D CD, 3D SIP and 3D Dextran. As dissociation behavior is more important in comparison with non-specific adsorption, slow dissociation rates lead to the saturation or permanent adsorption of analytes which cause complete destruction of platform. In results, fast dissociation of serum was recorded from 3D SIP and 3D Dextran when compared with 3D CD and 2D PEG surface which shows slow and almost same dissociation rates. The same behavior was also observed in cases of cell lysate adsorption and showing faster dissociation of cell lysate from SIP when compared with dextran. Results suggested the better performance of 3D SIP and 3D Dextran over other two platforms compared in this study.
- Non-specific adsorption of lysate and serum prior to carboxylation (a) sensorgram showing parallel response of non-specific adsorption of stem cell lysate and (b) human serum on PEG, CD, SIP and dextran surfaces.
3.4 SPRi analysis of adsorption of serum and cell lysate after carboxylation
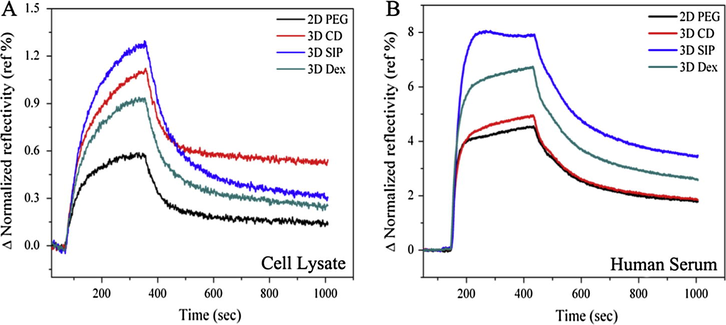
Carboxyl (—COOH) functional groups play an important role in the fabrication of most of the microarrays, so it is necessary and more important to check and compare the non-specific adsorption after carboxylation of hydroxyl (—OH) groups. After transformation of hydroxyl functional groups into carboxyl functional groups by 1:1 mixture of succinic anhydride and DMAP, activation and blocking steps and non-specific adsorption were then compared for all slides in same conditions and concentrations as explained above. Non-specific adsorptions of serum and cell lysate are shown in Fig. 5A and B. It is important to note that serum and cell lysate were adsorbed in high amount on carboxyl terminated surface as confirmed by the signal enhancement than hydroxy one due to more hydrophilic nature of surfaces. With high adsorption of serum and cell lysate after carboxylation, dissociation rates also became slower when compared with hydroxy terminated surfaces. In comparison, PEG and CD surfaces were performed poorly and in other hand, SIP and dextran surface seems more efficient to reduce non-specific adsorption when compared with former one, although CD shows better results when compared with PEG surface in terms of dissociation rates which is more faster in case of CD. Furthermore, signal to noise (SNR) ratio of non-specifically adsorbed serum and lysates was also calculated from each surface as shown in Fig. 6A and B.
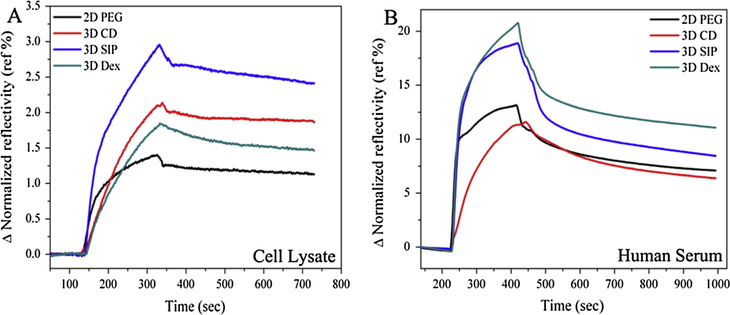
- Non-specific adsorption of cell lysate and serum after carboxylation (A) sensorgram showing parallel response of non-specific adsorption of stem cell lysate and (B) human serum on PEG, CD, SIP and dextran surfaces.
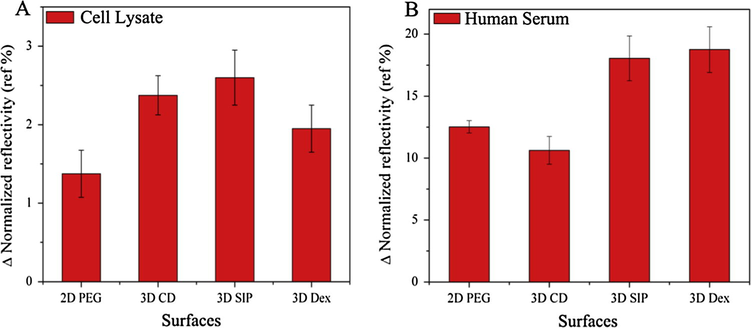
- Signal response on each surface, (A) graph representing the SPRi response for non-specific adsorption of cell lysate and (B) human serum onto PEG, CD, SIP and dextran surfaces with standard deviation.
3.5 MS analysis of adsorbed proteins from serum and cell lysate
After checked successfully by SPRi, the same slides were evaluated by MALDI–TOF analysis for identification of physically adsorbed lipids/proteins. For this purpose, the flow cells from slides were gently removed with the help of sharp forceps and then printing area was carefully cut with diamond cutters, and gently washed with diluted PBS and then with milli Q water in order to clean the surfaces. The outcome result of MALDI–TOF analysis suggested that instead of proteins, mainly lipids were non-specifically adsorbed in both cases i.e. cell lysate and serum. The identification of corresponding lipid/proteins was performed by database interrogation with the help of MASCOT search engine and LIPID maps (http://www.lipidmaps.org/). Obtained data from this search suggested the major adsorption of collagen α-1 protein, phosphatidylcholine and sphingomyelins lipid types. Interestingly, in both cases of cell lysate and serum, PEG and cyclodextrin surfaces showed high rate of non-specific adsorption when compared with SIP and dextran surfaces where only one or two types of lipid/protein were adsorbed (Figs. 7 and 8). After compared these results with those obtained from SPRi, It was easy to define the strong adsorption of lipid/proteins on PEG and dextran. Here, MS analysis results were in strong agreement with SPRi results. SIP and dextran surface produced best result overall in MS and SPRi analysis whether it showed only few proteins were adsorbed with very weak interaction and could be easily wiped off by buffer as proved by SPRi. The serum data were also showing the same tradition as cell lysate for all 4 surfaces except some difference in molecular weights of adsorbed protein/lipid because of their configuration. In whole, SIP and dextran proved to be best surface in terms of non-specific adsorption of cell lysate and serum in our study.
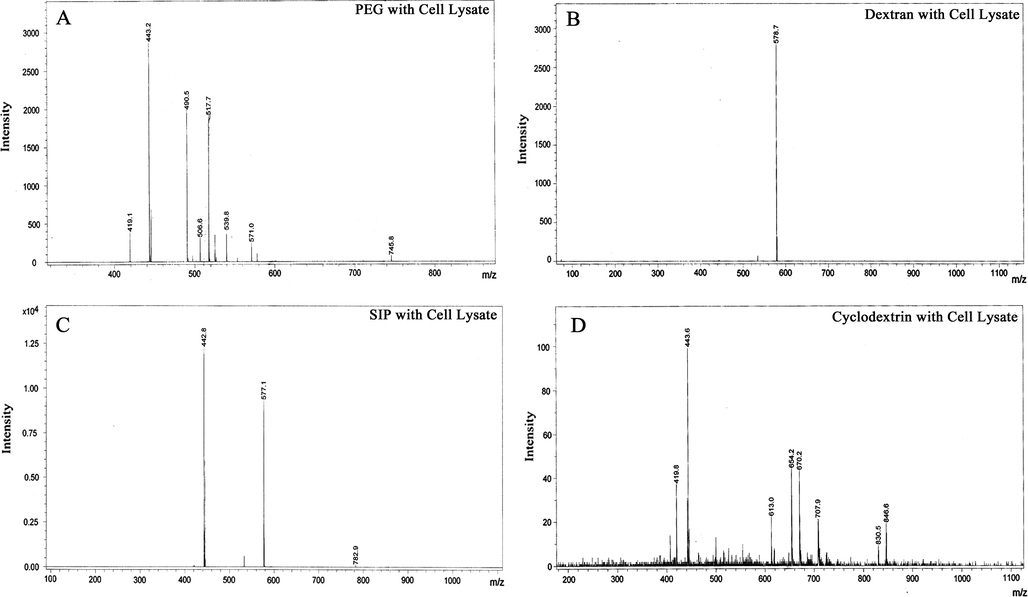
- MALDI–TOF characterization of adsorbed lipid/proteins from cell lysate onto (A) PEG, (B) dextran, (C) SIP and (D) cyclodextrin surfaces.
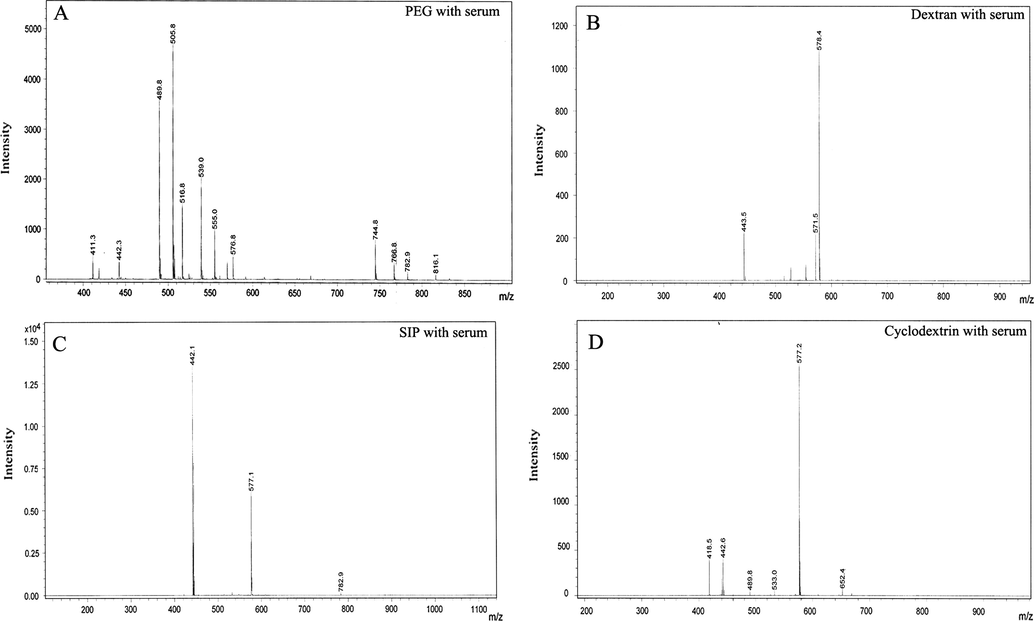
- MALDI–TOF characterization of adsorbed lipid/proteins from human serum onto (A) PEG, (B) dextran, (C) SIP and (D) cyclodextrin surfaces.
4 Discussion
This paper reports the comparative study of non-specific adsorption of human serum and induced pluripotent stem cell lysate on various advanced 3D biosensor surfaces by surface plasmon resonance imaging and mass spectrometry. Although, author reported these surfaces as non-fouling surface and have great applications in biosensors, but, previously these reported surfaces were validated only by using purified proteins and not complex system such as serum and cell lysate. Non-specific adsorption is a key problem in use of SPRi, especially for the drug discovery and biomarker study which always results into false positives and quenched the real and important information. For better understanding and selection of surface chemistries, here we demonstrated that all 3D platforms showed high non-specific adsorption especially after carboxylation step even more than 2D PEG surface. However, after high non-specific adsorption, fast dissociation rates were observed from the 3D SIP and 3D dextran surface when compared with 3D CD and 2D PEG assembled platforms which clearly showed the better performance of SIP and dextran surfaces. MALDI–TOF analysis of adsorbed proteins/lipid from cell lysate and serum was also performed to support the SPRi results. We conclude that, SIP and dextran structures can be further improved by structure modification and by trying different type of spacers such as methyl terminated polyethylene glycols instead of hydroxy one. Because hydroxy functional groups of spacer also changed to carboxyl during carboxylation of primary chains used for biomolecules immobilization in order to fabricate the microarray and lead to high non-specific adsorption, we strongly believe that the present study will surely help the researchers for further improvement in these reported surfaces so it can be applied in screening of complex biological structures such as, serum and cells.
Acknowledgments
We thank Prof. Jia Na for providing high quality mouse embryonic cell lysate and Prof. Hu Zhiyuan for providing human serum for this study. We gratefully acknowledge the financial support from 973 Program (2009CB930702) and NSFC Grant (61077064/60921001).
References
- Comparison of DNA, aminoethylglycyl PNA and pyrrolidinyl PNA as probes for detection of DNA hybridization using surface plasmon resonance technique. Biosens. Bioelectron.. 2010;25:1064-1069.
- [Google Scholar]
- Non-specific adsorption of crude cell lysate on surface plasmon resonance sensors. Langmuir. 2013;29:10141-10148.
- [Google Scholar]
- Modified peptide monolayer binding His-tagged biomolecules for small ligand screening with SPR biosensors. Analyst. 2011;136:3142-3148.
- [Google Scholar]
- Dry film refractive index as an important parameter for ultralow fouling surface coatings. Biomacromolecules. 2012;13:589-593.
- [Google Scholar]
- Methods of reducing non-specific adsorption in microfluidic biosensors. J. Micromech. Microeng.. 2010;20:075015.
- [Google Scholar]
- Development of a smart dynamic surface chemistry for surface plasmon resonance-based sensors for the detection of DNA molecules. J. Mater. Chem. B. 2013;1:5398-53402.
- [Google Scholar]
- Surface plasmon resonance-biosensor detects the diversity of responses against epidermal growth factor in various carcinoma cell lines. Biosens. Bioelectron.. 2012;32:202-207.
- [Google Scholar]
- Label-free analysis of biomolecular interactions using SPR imaging. Biotechniques. 2011;50:32-40.
- [Google Scholar]
- Label-free detection of proteins in crude cell lysate with antibody arrays by a surface plasmon resonance imaging technique. Anal. Chem.. 2005;77:7115-7121.
- [Google Scholar]
- Direct detection of carcinoembryonic antigen autoantibodies in clinical human serum samples using a surface plasmon resonance sensor. Colloids Surf. B. 2009;70:1-6.
- [Google Scholar]
- Real-time monitoring biomarker expression of carcinoma cells by surface plasmon resonance biosensors. Chem. Commun.. 2012;48:10389-10391.
- [Google Scholar]
- A novel hydrogel matrix on gold surfaces in surface plasmon resonance sensors for fast and efficient covalent immobilization of ligands. J. Chem. Soc. Chem. Commun. 1990:1526-1528.
- [Google Scholar]
- Surface initiated polymerization from substrates of low initiator density and its applications in biosensors. ACS Appl. Mater. Interf.. 2010;2:3223-3230.
- [Google Scholar]
- Biochip functionalization using electrowetting-on-dielectric digital microfluidics for surface plasmon resonance imaging detection of DNA hybridization. Biosens. Bioelectron.. 2009;24:2218-2224.
- [Google Scholar]
- Multi-analyte SPR immunoassays for environmental biosensing of pesticides. Anal. Bioanal. Chem.. 2007;387:1449-1458.
- [Google Scholar]
- Glycoprotein profiling of stem cells using lectin microarray based on surface plasmon resonance imaging. Anal. Biochem. 2014
- [CrossRef] [Google Scholar]
- High-throughput SPR sensor for food safety. Biosens. Bioelectron.. 2009;24:1399-1404.
- [Google Scholar]
- Dendrimer functionalization of gold surface improves the measurement of protein–DNA interactions by surface plasmon resonance imaging. Biosens. Bioelectron.. 2013;43:148-154.
- [Google Scholar]
- Properties of ionic liquids on Au surfaces: non-conventional anion exchange reactions with carbonate. Chem. Commun.. 2011;47:10644-10646.
- [Google Scholar]
- Design of surface modifications for nanoscale sensor applications. Sensors. 2015;15:1635-1675.
- [Google Scholar]
- Surface plasmon resonance imaging for nucleic acid detection. Anal. Bioanal. Chem.. 2013;405:573-584.
- [Google Scholar]
- Substrate independent approach for the generation of functional protein resistant surfaces. Biomacromolecules. 2011;12:1058-1066.
- [Google Scholar]
- Development of surface plasmon resonance imaging biosensors for detection of ubiquitin carboxyl-terminal hydrolase L1. Anal. Biochem.. 2015;469:4-11.
- [Google Scholar]
- Design of a universal biointerface for sensitive, selective, and multiplex detection of biomarkers using surface plasmon resonance imaging. Analyst. 2013;138:6052-6062.
- [Google Scholar]
- 3D small molecule microarray with enhanced sensitivity and immobilization capacity monitored by surface plasmon resonance imaging. RSC Adv. 2014
- [CrossRef] [Google Scholar]
- Small molecule microarray screening methodology based on surface plasmon resonance imaging. Arab. J. Chem.. 2019;12:2111-2117.
- [CrossRef] [Google Scholar]
- Detection of carcinoembryonic antigens using a surface plasmon resonance biosensor. Sensors. 2008;8:4282-4295.
- [Google Scholar]
- Label-free immunosensing for α-fetoprotein in human plasma using surface plasmon resonance. Anal. Biochem.. 2007;365:201-207.
- [Google Scholar]
- Ultralow fouling and functionalizable surface chemistry based on a zwitterionic polymer enabling sensitive and specific protein detection in undiluted blood plasma. Anal. Chem.. 2008;80:7894-7901.
- [Google Scholar]
- Detection of picomolar levels of interleukin-8 in human saliva by SPR. Lab Chip. 2005;5:1017-1023.
- [Google Scholar]







