ssDNA-QDs/GO multicolor fluorescence system for synchronous screening of hepatitis virus DNA
⁎Corresponding author. jjyang2009@126.com (Jiajia Yang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Viral hepatitis is a common infectious disease caused by five viruses (hepatitis virus A, B, C, D, and E). Given the diversity of hepatitis virus, rapid screening and accurate typing of viral hepatitis are the prerequisites for hepatitis therapy. Here, a multicolor fluorescence system was constructed by combining with the multi-color fluorescence properties of CdSe/ZnS quantum dots (QDs, emission wavelengths: 525 nm, 585 nm and 632 nm) and the broad-spectrum fluorescence quenching performance of GO. Taking advantage of the specific recognition of ssDNA modified CdSe/ZnS QDs to target hepatitis virus DNA, the constructed system could effectively distinguish hepatitis A virus DNA (HAV-DNA), hepatitis B virus DNA (HBV-DNA), and hepatitis C virus DNA (HCV-DNA) in a homogeneous solution. Based on the different adsorption property of GO for ssDNA and dsDNA, the fluorescence Forster resonance energy transfer (FRET) process between ssDNA modified QDs and GO could be regulated. The fluorescence signal of the constructed system presented a sensitive response to HAV-DNA, HBV-DNA, and HCV-DNA content in the range of 1.0–192 nM, 8.0–192 nM, and 1.0–128 nM, respectively. The limit of detection for HAV-DNA, HBV-DNA, and HCV-DNA is 0.46 nM, 1.53 nM, and 0.58 nM. The constructed system can be used to screen hepatitis virus DNA in real samples, which provides an alternative strategy for rapid screening and diagnosis of viral hepatitis.
Keywords
Multicolor quantum dot
Synchronous screening
Graphene oxide
Hepatitis virus DNA
- QDs
-
CdSe/ZnS quantum dots
- HAV-DNA
-
Hepatitis A virus DNA
- HBV-DNA
-
Hepatitis B virus DNA
- HCV-DNA
-
Hepatitis C virus DNA
- QDs@CHAV-DNA
-
Complementary ssDNA strands of hepatitis A virus DNA modified QDs
- QDs@CHBV-DNA
-
Complementary ssDNA strands of hepatitis B virus DNA modified QDs
- QDs@CHCV-DNA
-
Complementary ssDNA strands of hepatitis C virus DNA modified QDs
Abbreviations
1 Introduction
Hepatitis defined as the inflammation of the liver tissue is a globally common disease. Although there are many pathogenic factors that can induce hepatitis (viruses, bacteria, parasites, chemical poisons, drugs, alcohol, and autoimmune factors), viral hepatitis remains the predominant cause for liver disease and has been a global public health problem due to its strong contagiousness, complex transmission route and high incidence (Semaine et al., 2006; van Leeuwen et al., 2022). According to statistics, approximately 1.3 million people die as a result of viral hepatitis each year worldwide, which is mainly attributed to cirrhosis and liver cancer caused by hepatitis B virus (HBV), hepatitis C virus (HCV), hepatitis D virus (HDV), acute infection of hepatitis A virus (HAV) and liver failure caused by hepatitis E virus (HEV) (Krugman et al., 1997). Given the diversity of viral hepatitis types and the limited cross-immunization between them, humans can be co-infected, or overlapping infected with hepatitis viruses. Thus, diagnosis and classification of viral hepatitis is still a challenging topic.
Viral hepatitis can be diagnosed using serology tests (antibody) or molecular (presence of viral RNA or DNA) diagnostics. Hepatitis virus DNA detection is an effective method with high assay efficiency and low sample requirement. Up to now, a large number of DNA detection methods have been developed for the diagnosis of viral hepatitis including high performance liquid chromatography(Wolford et al., 2000), real-time polymerase chain reaction (PCR) (Geng et al., 2006; Wang et al., 2013b), surface-enhanced Raman scattering(Zhu et al., 2021), electrochemical detection(Manzano et al., 2018; Wang et al., 2013a; Wang et al., 2013b), and colorimetric detection(Mao et al., 2016), molecularly imprinted polymers-RLS sensor, resonance light scattering sensor, molecular imprinting fluorescence sensor (Luo, et al., 2016), gas-responsive molecularly imprinted sensor. These strategies are highly sensitive, but most of them are employed to detect a single type of hepatitis virus DNA. Accordingly, it is difficult to achieve rapid typing of hepatitis types. Due to the limited information provided by single target analysis, false-positive results are inevitable, which will result in a relatively high rate of misdiagnosis and missed diagnosis (He et al., 2017). In order to overcome these shortcomings, multiplexed target DNA detection will be an ideal approach for accurate diagnosis of viral hepatitis.
Compared with single detection, multi-biomarkers simultaneous analyses are becoming increasingly popular, which enable to detect two or more targets in a single analyze with high detection flux, low sample requirement, shorter assay time, and low expenditure(Liu et al., 2016b). Currently, several materials have been used for simultaneous detection, such as organic fluorescence dyes (Ren et al., 2022), quantum dots (QDs) (Hu et al., 2021; Jie et al., 2017; Li et al., 2020), metal nanoparticles (Kim & Jurng, 2011; Zhang et al., 2020), and so on. Although organic fluorescent dyes have been successfully applied to the simultaneous detection of multiple targets, the simultaneous detection of multiple targets requires multiple excitation operations as the inconsistent excitation wavelength of each organic fluorescent dye restricts them from being excited synchronously at the same excitation wavelength. Compared with fluorescent dyes, the multicolor fluorescence properties of QDs under single excitation wavelength endow them with excellent flexibility in developing synchronous detection system (Chandan et al., 2018; Shen et al., 2021; Xiang et al., 2020). Especially, QDs can be assembled with a variety of nano-materials to form composite detection systems (Adegoke & Forbes, 2016; Miao et al., 2016). Typically, QDs/GO composite system can be served as an excellent platform for target detection based on fluorescence resonance energy transfer (FRET), wherein QDs is used as energy donor and GO acts as energy acceptor (Zhang et al., 2011). When QDs and GO are close to each other, the energy will transfer from the QDs to the GO and then the fluorescence signal of QDs will be quenched (Zhang et al., 2011; Arvand & Mirroshandel, 2019). Therefore, promoting or preventing the interaction between QDs and GO by the mediation of targets has been used as an ingenious strategy for the construction of various probes (Arvand & Mirroshandel, 2019). Numerous studies have demonstrated that GO can strongly adsorb single-stranded DNA (ssDNA) via hydrophobic and π-π stacking interactions between DNA nucleobases and sp2-hybridized atoms of GO nanosheets, but it has low affinity toward double-stranded DNA (dsDNA)(Liu et al., 2016a; Wang et al., 2018). Based on these unique properties of GO (Park et al., 2013; Wu et al., 2011), several ssDNA modified QDs/GO fluorescent probes have been reported (Arvand & Mirroshandel, 2019; Li et al., 2017). These studies provided valuable references for accurate detection targets in complex matrices. However, they mainly focus on the detection of single target. Combined with our previous work (Yang et al., 2018) and inspired by the research of Xiang et al. (Xiang et al., 2020), the construction of QDs/GO composite fluorescence system may be an effective approach to realize the synchronous screening of multiple viral hepatitis DNA.
In this paper, a multicolor fluorescence system was attempted to construct for synchronous screening of hepatitis virus DNA. As illustrated in Scheme 1, three water-soluble CdSe/ZnS QDs with different maximum emission wavelength at 525 nm, 585 nm and 632 nm were firstly modify by using of complementary ssDNA strands of HAV-DNA, HBV-DNA, and HCV-DNA though cross-linking reaction. Taking advantage of the multicolor fluorescence property of CdSe/ZnS QDs and unique quenching performance of GO, a multicolor fluorescence response system was constructed, in which QDs would be adsorbed on the surface of GO by the mediation ssDNA ligands, and the fluorescence signals of QDs would be synchronously quenched through FRET. When the HAV-DNA, HBV-DNA, and HCV-DNA were captured by complementary ssDNA ligands modified QDs through hybridization reaction, FRET will be blocked due to weak adsorption of GO to double-stranded DNA (dsDNA), and strong fluorescence signals will be detected. Compared with single fluorescence and double-color fluorescence detection, a multicolor fluorescence system was proposed by virtue of QDs and GO, which is expected to serve as an alternative strategy for rapid screening and diagnosis of viral hepatitis.
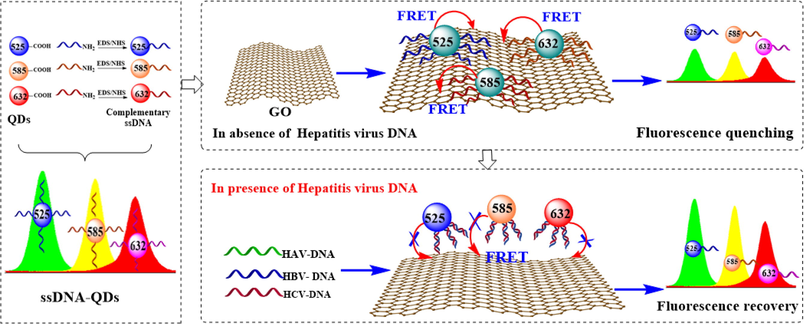
- Schematic illustration of ssDNA-QDs/GO multicolor fluorescence system for synchronous screening of hepatitis virus DNA.
2 Experimental section
2.1 Materials and chemicals
The DNA sequences (shown in Table S1) used in the experiment were synthesized and purified by Sangon Biotechnology Co. ltd. (Shanghai, China). N-Hydroxylsuccinimide sodium salt (NHS) and 1-ethyl-3-(3-(dimethylamino) propyl) carbodiimide hydrochloride (EDC) were purchased from Sigma-Aldrich (St. Louis, USA). Three PEG-capped and carboxyl-modified CdSe/ZnS QDs (the maximum emission wavelength at 525 nm, 585 nm and 632 nm, the excitation wavelength at 340 nm) were provided by Suzhou Xingshuo Nanotech Co., ltd. (Suzhou, China). GO was purchased from Nanjing Xianfeng Nanomaterials Technology Co., ltd. (Nanjing, China). The other reagents used were of analytical grade, and all the solutions were prepared with twice-distilled water.
2.2 Instruments
The morphologies of all samples were characterized by high-resolution transmission electron microscope (HR-TEM) (JEM-2100, Hitach, Japan). Fourier transform infrared spectroscopy was performed on a Magna-560 spectrometer (Nicolet, Madison, WI). The UV–vis absorption spectra were monitored using a Shimadzu UV-29100 spectrophotometer (Tokyo, Japan). All fluorescence spectra were measured on a Cary Eclipse fluorescence spectrophotometer (Varian American Pty ltd., USA) with a 1 cm path-length quartz cell. The excitation wavelength was 340 nm, and the silt widths of excitation and emission were set at 10 nm.
2.3 Preparation of ssDNA modified QDs
Three ssDNA modified QDs were prepared as follows: 23.0 μL EDC (10 mg mL−1) and 40 μL QDs (10 μmol/L) were added to 200 μL PBS buffer (100 mmol/L, pH = 6.0). The solution was continuously and gently agitated for 30 min to activate the carboxyl group. Subsequently, the pH of the above solution was adjusted immediately to 8.0 with PBS (pH = 8.5) prior to the addition of 13.8 µL NHS (10 mg mL−1) and a certain concentration of complementary DNA. The mole ratio of complementary DNA to QDs was 40:1. After gently shaking for 3 h at room temperature, the resulting solutions were placed in a refrigerator at 4 ℃ overnight. Then, an ultrafilter tube (30 kD) was used to centrifuge the solution to remove excess ssDNA, EDC, and NHS. The final products were diluted to 5 mL for the subsequent experiments.
2.4 Synchronous screening of hepatitis virus DNA
The as-prepared QDs@CHAV-DNA, QDs@CHBV-DNA, and QDs@CHCV-DNA solution with the same concentration was mixed together in equal proportions. The mixed solution was abbreviated as QDs@CDNA. Then, 60 µL QDs@CDNA mixed solution and 500 µL Tris-HCL buffer (20 mmol/L Tris-HCL, 0.1 mol/L NaCl, pH = 7.4) were added to a series of 5.0 mL colorimeter tubes. And then HAV-DNA, HBV-DNA and HCV-DNA solution with different concentrations were added and a constant volume of 1 mL was achieved using distilled water. After mixed thoroughly and hybridized for 60 min in a thermostat water bath at 37 ℃, 150 µL GO (0.5 mg mL−1) was added into the above mixed solution, and the solution volume was diluted to 2.5 mL with distilled water and incubated for 30 min at room temperature. The fluorescence spectra were recorded from 400 nm to 700 nm at an excitation wavelength of 340 nm, and the slit widths for excitation and emission were set at 10 nm.
2.5 Screening of hepatitis virus DNA in real sample
The serum samples were purchased from Sigma-Aldrich Company, which were diluted 10 times with distilled water before use. Then,16 nM and 64 nM of the target hepatitis virus DNA were added to the samples, respectively. The labeled samples with different standard additions were examined at an excitation wavelength of 340 nm under the optimized conditions, and the fluorescence spectra of the QDs@CDNA and GO/DNA system were observed in the range of 400–700 nm.
3 Results and discussion
3.1 Construction of multicolor fluorescence system
Three kinds of QDs have good dispersion and spherical morphology (Fig. S1A-C). The average hydrodynamic size of QDs(5 2 5), QDs(5 8 5), and QDs(6 3 2) is 11.62 nm, 12.54 nm, and 13.44 nm, respectively (Fig. S1D-F). In order to verify the feasibility of constructing multicolor fluorescence system using QDs with different maximum emission wavelength, the fluorescence emission spectrums of QDs(5 2 5), QDs(5 8 5), and QDs(6 3 2) were firstly measured with the same concentration at the exciting wavelength of 340 nm. As presented in Fig. 1A, the maximum emission intensities of QDs(5 2 5), QDs(5 8 5), and QDs(6 3 2) corresponded to 525 nm, 585 nm, and 632 nm, respectively. It is consistent with the instruction manual provided by Suzhou Xingshuo Nanotech Co., ltd, and can be found on the company website. Besides, it also could be found that there is no obvious overlap between the emission spectrum of three QDs, which indicated that the emission signal of each QDs can be detected individually in a mixed system of QDs(5 2 5), QDs(5 8 5), and QDs(6 3 2). Moreover, it was exciting that no significant change was observed in the fluorescence intensity of QDs(5 2 5) after QDs(5 8 5) and QDs(6 3 2) were added in succession, and the emission spectrum of QDs(5 2 5) did not show obvious blue-shift and red-shift (Fig. 1B). The similar results were also observed even if the sampling sequence of the QDs was swapped (Fig. 1C, D).
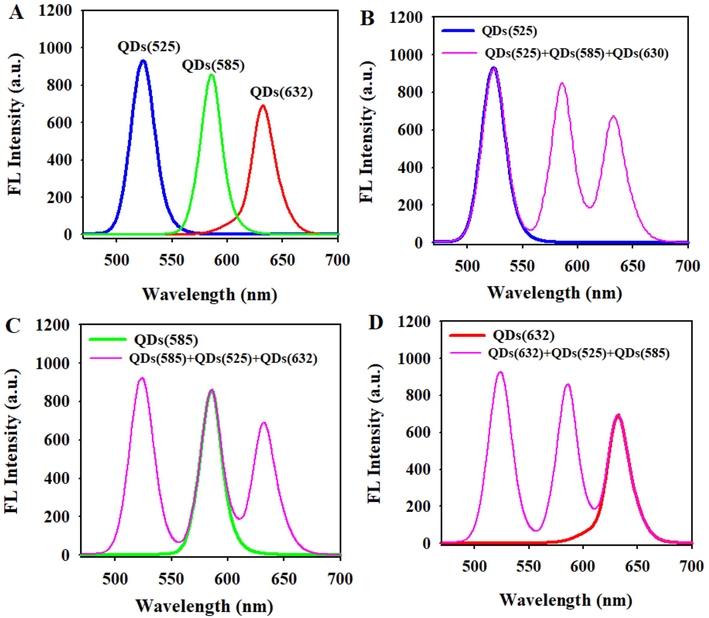
- (A)The emission fluorescence spectra of pure QDs(5 2 5), QDs(5 8 5) and QDs(6 3 2). (B)-(D) Comparison of the fluorescence intensity of a single QDs and that of mixture QDs.
In order to realize the selective recognition of three hepatitis viral DNA, QDs(5 2 5), QDs(5 8 5), and QDs(6 3 2) was modified by complementary ssDNA strands of HAV-DNA, HBV-DNA and HCV-DNA, respectively. The modified QDs were referred to as QDs@CHAV-DNA, QDs@CHBV-DNA, and QDs@CHCV-DNA. Compared with the unmodified QDs, typical DNA absorption peaks between 260 and 280 nm can be observed in UV–vis absorption spectrums of the QDs@CHAV-DNA, QDs@CHBV-DNA, and QDs@CHCV-DNA (Fig. S2), indicating the ssDNA strands were successfully modified on the surface of QDs. As shown in Fig. 2A, the fluorescence intensity of modified QDs did not change obviously relative to those of unmodified QDs. The solution of QDs@CHAV-DNA, QDs@CHBV-DNA, and QDs@CHCV-DNA presented green, yellow and pink fluorescence under UV illumination, respectively (Fig. 2B). The kinetic analysis of the change of fluorescence intensity of three modified QDs showed that the fluorescence intensity of QDs@CHAV-DNA, QDs@CHBV-DNA, and QDs@CHCV-DNA remained relatively stable within 60 min (Fig. S3A-C). The fluorescence spectrums of QDs@CDNA remained almost unchanged (inset in Fig. S3 A-C). The quantum yield of QDs(5 2 5), QDs(5 8 5), and QDs(6 3 2) was 83.4 %, 87.3 %, 89.5 %, respectively. After modification with HAV-DNA, HBV-DNA, and HCV-DNA, the quantum yield of QDs@CHAV-DNA, QDs@CHBV-DNA, and QDs@CHCV-DNA was 80.1 %, 83.1 %, 84.6 %, respectively. It indicated that ssDNA modification does not change the fluorescence properties of QDs(5 2 5), QDs(5 8 5), and QDs(6 3 2). These results indicated that the QDs@CHAV-DNA, QDs@CHBV-DNA, and QDs@CHCV-DNA have strong fluorescence properties and good optical stability.
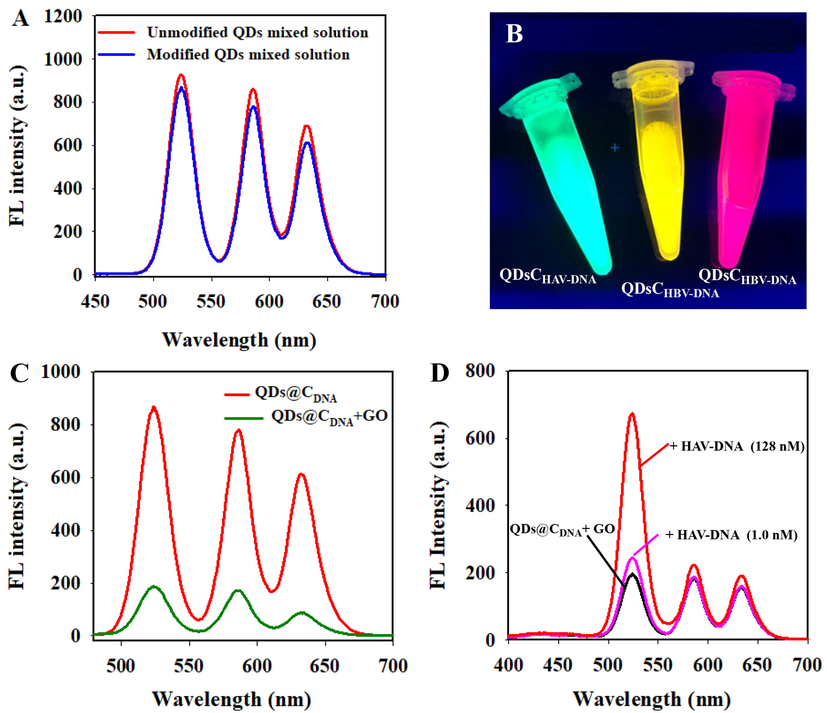
- (A)The emission fluorescence spectra of the modified QDs and the unmodified QDs. (B) Photograph of the QDs@CHAV-DNA, QDs@CHBV-DNA, and QDs@CHCV-DNA under UV illumination. (C) The change of fluorescence emission spectra of QDs@CDNA in presence of GO (30 μg mL−1). (D) Fluorescence emission spectra of QDs@CDNA/GO with different concentrations of HAV-DNA (1.0 nM and 128 nM).
As aforementioned, GO can effectively adsorb ssDNA via hydrophobic and π-π stacking interactions between DNA nucleobases and sp2-hybridized atoms of GO nanosheets, but it has low affinity toward double-strand DNA (dsDNA) (Liu et al., 2016a; Wang et al., 2018). Besides, GO can effectively quench the fluorophore of QDs through energy resonance transfer. Here, one of our concerns is that whether the fluorescence of QDs with three emission wavelengths can be synchronously quenched. Consequently, the fluorescence quenching properties of GO for QDs@CHAV-DNA, QDs@CHBV-DNA, and QDs@CHCV-DNA were investigated. From Fig. S4 it could be seen that the GO is transparent due its single layered structure and has irregular edges. As illustrated in Fig. 2C, the fluorescence intensity of the mixed system significantly reduced after the addition of GO, and the fluorescence quenching efficiency for QDs@CHAV, QDs@CHBV and QDs@CHCV was approximately 79.71 %, 78.04 %, and 84.56 %, respectively. Thus, the fluorescence of QDs@CHAV, QDs@CHBV and QDs@CHCV could be quenched simultaneously by GO.
As shown in Fig. 2D, when HAV-DNA was incubated with QDs@CHAV-DNA for 60 min prior to the addition of GO, the fluorescence intensity of QDs@CHAV-DNA was obviously recovered at 525 nm, but there was no obvious change of the fluorescence intensity at 585 nm and 632 nm. If the HBV-DNA or HCV-DNA was incubated with QDs@CHBV-DNA or QDs@CHCV-DNA before adding GO, the fluorescence intensity of the QDs@CHBV-DNA and QDs@CHCV-DNA at 585 nm, and 632 nm also recovered obviously, illustrating the proposed system can effectively recognize and distinguish three kinds of target DNA(Fig. S5A-B). If the DNAHAV, DNAHBV, and DNAHCV were mixed in equal proportion and incubated with QDs@CDNA together, the fluorescence intensity was recovered simultaneously at 525 nm, 585 nm and 632 nm (Fig. S5C). This further confirmed that current conception is feasible and can be used for constructing a synchronous screening system for three kinds of hepatitis virus DNA.
3.2 Optimization of conditions for synchronous screening of hepatitis viral DNA
The effect of GO concentration on the fluorescence intensity of the multicolor fluorescence system was investigated. As shown in Fig. S6A, the fluorescence intensity corresponding to 525 nm, 585 nm and 632 nm decreased gradually with the increase of GO concentration. From the response curve of the fluorescence intensity ratio (F/F0) to GO concentration (where F and F0 refer to the fluorescence intensity of the multicolor system in the presence and absence of GO, respectively), it could be seen that the fluorescence intensity did not change when the GO concentration was higher than 30 μg mL−1 (Fig. S6 B). Thus, 30 μg mL−1 GO was selected for the subsequent experiments.
As we know, weak alkaline medium is favorable for DNA hybridization. The effect of pH on fluorescence intensity of the multicolor fluorescence system was investigated over the pH range of 6.0–8.5 (Fig. S6C). The fluorescence quenching efficiency of GO for three kinds of QDs@CDNA was relatively high at pH 7.4, but decreased at pH higher than 7.4. In the presence of target DNA, the maximum fluorescence recovery ratio was achieved at pH = 7.4. In order to ensure the high sensitivity of the system, Tris-HCl buffer with pH 7.4 was selected for the subsequent experiments.
Fig. S6D displayed the effects of incubation time on analytical performance of the multicolor fluorescence system. The fluorescence intensity of the system in the presence of GO gradually decreased with the prolonged incubation time and plateaued within 30 min at room temperature. Thus, 30 min was selected as the reaction time between QDs@CDNA and GO. The hybridization reaction time of QDs@CDNA/GO and target DNAs was further optimized. It could be seen that the fluorescence intensity ratio of the system gradually increased with the extension of hybridization time, and did not obviously change after 60 min (Fig. S6E). Thus, 60 min was selected as the hybridization time.
The ionic strength played an important role in DNA hybridization reaction. the fluorescence recovery ratio F/F0 was optimal when the NaCl concentration in hybridized buffer was 50 mM (Fig. S6F). Therefore, 50 mM NaCl solution was used in this study.
3.3 Synchronous screening of hepatitis viral DNA
The analytical performance of the constructed system was evaluated under the optimized conditions by adding three kinds of hepatitis viral DNA with the same proportion. As shown in Fig. 3A, the fluorescence intensities of the system at 525 nm, 585 nm, and 632 nm simultaneously increased with the increase of HAV-DNA, HBV-DNA, and HCV-DNA concentration. Fig. 3B showed a good linear relationship between the fluorescence intensity ratio F/F0 (F and F0 refer to the fluorescence intensity of the system in the presence and absence of target DNA, respectively) and the concentration of three hepatitis virus DNA. The F/F0 presented a sensitive response to the concentration of HAV-DNA, HBV-DNA, and HCV-DNA in the range of 1.0–192 nM, 8.0–192 nM, and 1.0–128 nM, respectively. The standard curves for HAV-DNA, HBV-DNA and HCV-DNA were F/F0 = 0.012C(HAV-DNA) + 1.19 (R2 = 0.992), F/F0 = 0.0102C(HBV-DNA) + 1.27(R2 = 0.984) and F/F0 = 0.0132C(HCV-DNA) + 1.25 (R2 = 0.985), respectively. Based on 3σ/S, the detection limit for HAV-DNA, HBV-DNA, and HCV-DNA were calculated as 0.46 nM, 1.53 nM, and 0.58 nM. The relative standard deviation (RSD) for five times repeated detection of 32 nM HAV-NDA, HBV-DNA, and HCV-DNA was about 3.07 %, 3.73 %, 3.45 %, respectively. It indicated acceptable reproducibility of the proposed multicolor fluorescence system.
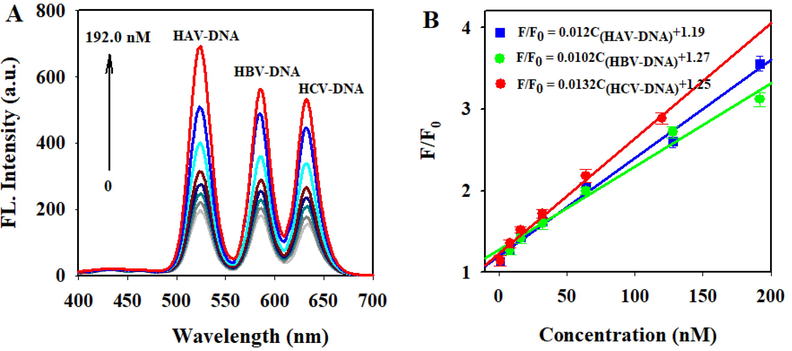
- (A) Fluorescence spectra of the multicolor fluorescence system with different concentrations of HAV-DNA, HBV-DNA, and HCV-DNA (0, 1.0, 8.0, 16.0, 32.0, 64.0, 128.0, 192.0 nM). (B)The relationships between fluorescence intensity ratio (F/F0) and the concentration of HAV-DNA, HBV-DNA, and HCV-DNA.
Table 1 shows the comparison between the proposed method and other reported methods for hepatitis virus DNA in terms of detection range and limit. The analytical parameters of the constructed system are comparable to or better than those of fluorescence methods (Chen et al., 2015; Zhu et al., 2015; Zeng et al., 2018; Fakih et al., 2017). Compared with single or double target detection, the constructed probe could rapidly detect three kinds of hepatitis viral DNA simultaneously. Although the sensitivity of our constructed system is inferior to those of some electrochemical methods and molecularly imprinted fluorescence sensor (Honorato Castro et al., 2014; Liu et al., 2016b; Luo et al., 2019), modifier preparation and electrode modification in these strategies are sophisticated and time consuming. In term of the operation process, the proposed system is convenient and time-efficient.
| Method | Material | Target DNA | Linear range | Detection limit | Ref. |
|---|---|---|---|---|---|
| Fluorescence polarization | SiO2 NP–DNA/Ag NCs | HBV | 1–200 nM | 0.65 nM | Chen et al., 2015 |
| Fluorescence detection | NH2-UCNPs and Au NP | HBV | 0–50 nM | 0.25 nM | Zhu et al., 2015 |
| Fluorescence detection | Fluorescent dye | HCV | 50–400 nM | 24.57 nM | Zeng et al., 2018 |
| Fluorescence detection | Gold nanoparticles-coated polystyrene beads | HBV | — | 0.65 nM | Fakih et al., 2017 |
| Fluorescence detection | QDs and magnetic nanospheres | HBV HCV |
0.5–10 nM 0.5–8.0 nM |
0.148 nM 0.077 nM |
Hu et al., 2013 |
| Electrochemical detection | Graphite electrodes | HBV | 0.18–1.8 μM | 0.18 μM | Honorato Castro et al., 2014 |
| Molecularly imprinted fluorescence sensor | HAV | 0.3–95 nM | 3.4 PM | Luo, et al., 2016 | |
| HBV | 0.3–90 nM | 5.3 PM | |||
| Electrochemical detection | QDs and Au nanoparticles | HBV HCV |
0.0005–0.5 nM 0.001–1 nM |
0.08 pM 0.34 pM | Liu et al., 2016b |
| Fluorescence detection | ssDNA-QDs/GO | HAV HBV HCV |
1.0–192 nM 8.0–192 nM 1.0–128 nM |
0.46 nM 1.53 nM 0.58 nM |
This work |
3.4 Specificity
The specific performance of the constructed multicolor fluorescence system was investigated by recognition of three hepatitis virus DNA in a coexistence system containing their single-base, double-base, and triple-base mismatched ssDNA strands. As displayed in Fig. 4, the fluorescence intensity of three hepatitis virus DNA was apparently recovered at 525 nm, 585 nm and 632 nm, which corresponded to high fluorescence intensity ratio (F/F0). Compared with hepatitis virus DNA, their single-base, double-base, and triple-base mismatched ssDNA strands could not induce significantly recovery of the fluorescence. This is mainly due to the low hybridization capability of ssDNA towards its base mismatched complementary sequences. The constructed system can effectively distinguish and screen three kinds of hepatitis virus DNA from numerous ssDNA strands. To evaluate the anti-interference of the proposed multicolor fluorescence system, the effect of some coexistent substances (eg. biomolecules and ions) in the biological fluids on the detection of hepatitis virus DNA was investigated. As displayed in Table S2, 100 μM Na+, K+, and Ca2+ can hardly affect the detection of three hepatitis virus DNA. Moreover, 200 μM l-Cys, l-Gly, ATP, and BSA also had no obvious effect on the detection results. All of the observations above clearly indicated the high selectivity of the proposed method.
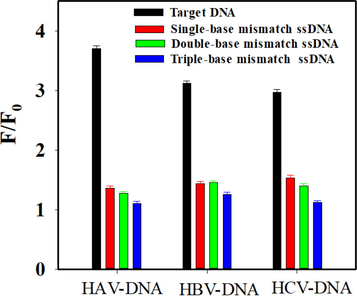
- Selectivity investigation of the proposed method in the presence of target DNA and different base mismatch strands. The concentration of HAV-DNA, HBV-DNA, and HCV-DNA was 192 nM, 192 nM and 128 nM, respectively. The concentration of GO was 30 μg mL−1. The concentration of base mismatched ssDNA was the same as the target DNA.
3.5 Screening of hepatitis viral DNA in serum sample
In order to investigate the applicability of the proposed system, spiked recovery experiments were performed by adding three kinds of hepatitis virus DNA with two concentrations into the diluted serum sample. The results were presented in Table 2. The recoveries for HAV-DNA, HBV-DNA and HCV-DNA were 92.19 %-108.20 %, 94.13 %-105.13 %, and 91.18 %-94.56 %. These results demonstrated the potential applicability of the proposed system for diagnosis of viral hepatitis by screening viral DNA.
| Hepatitis virus DNA | Serum samples (nM) | Added amount (nM) | Detected (nM) (Mean ± SD) | Recovery ratio (%) | RSD (%) |
|---|---|---|---|---|---|
| HAV-DNA | Not detected | 16.0 | 14.75 ± 0.64 | 92.19 | 4.03 |
| 64.0 | 69.25 ± 3.02 | 108.20 | 4.71 | ||
| HBV-DNA | Not detected | 16.0 | 15.06 ± 0.57 | 94.13 | 3.56 |
| 64.0 | 67.28 ± 2.74 | 105.13 | 4.29 | ||
| HCV-DNA | Not detected | 16.0 | 15.13 ± 0.62 | 94.56 | 3.89 |
| 64.0 | 58.87 ± 3.03 | 91.98 | 4.74 |
4 Conclusion
By virtue of the multi-color fluorescence properties of QDs, as well as the unique properties of GO including selective adsorption of ssDNA and efficient fluorescence quenching properties, ssDNA-QDs/GO multicolor fluorescence system was constructed for synchronous screening of DNA sequences originating from different types of hepatitis virus. Taking HAV-DNA, HBV-DNA and HCV-DNA as models, the constructed system displayed desirable performance for discriminating and recognizing three kinds of viral hepatitis DNA. Moreover, synchronous detection of HAV-DNA, HBV-DNA and HCV-DNA could be achieved by measuring the change of fluorescence signals of the system under a single wavelength excitation. The proposed strategy can serve as an alternative method for rapid screening of viral hepatitis through simple steps. In addition, the present work also provides a certain reference for the high-throughput detection of multiple disease markers through multi-color QDs and GO or other nanomaterials.
Author contributions
All authors contributed to literature research and manuscript writing.
Acknowledgements
This work was supported by the Natural Science Foundation of Shanxi Province of China (NO. 201901D211390) and Natural Science Foundation of Shanxi Normal University.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- l-Cysteine-capped core/shell/shell quantum dot–graphene oxide nanocomposite fluorescence probe for polycyclic aromatic hydrocarbon detection. Talanta. 2016;146:780-788.
- [Google Scholar]
- An efficient fluorescence resonance energy transfer system from quantum dots to graphene oxide nano sheets: application in a photoluminescence aptasensing probe for the sensitive detection of diazinon. Food Chem.. 2019;280:115-122.
- [Google Scholar]
- Quantum dots as fluorescent probes: synthesis, surface chemistry, energy transfer mechanisms, and applications. Sens. Actuators B. 2018;258:1191-1214.
- [Google Scholar]
- A SiO2 NP–DNA/silver nanocluster sandwich structure-enhanced fluorescence polarization biosensor for amplified detection of hepatitis B virus DNA. J. Mater. Chem. B. 2015;3(6):964-967.
- [Google Scholar]
- Gold nanoparticles-coated polystyrene beads for the multiplex detection of viral DNA. Sens. Actuators B. 2017;250:446-452.
- [Google Scholar]
- Geng, H., Hua, B., Wang, H., Cao, Y., Sun, Y., Yu, A.J.J.o.V.M. 2006. Dual-probe assay for detection of lamivudine-resistance hepatitis B virus by real-time PCR. Journal of Virological Methods, 132(1-2), 25-31
- Multiplexed photoluminescent sensors: towards improved disease diagnostics. Chem. Soc. Rev.. 2017;46(22):6687-6696.
- [Google Scholar]
- Preparation of genosensor for detection of specific DNA sequence of the hepatitis B virus. Appl. Surf. Sci.. 2014;314:273-279.
- [Google Scholar]
- DNA functionalized double quantum dots-based fluorescence biosensor for one-step simultaneous detection of multiple microRNAs. Talanta. 2021;235:122763
- [Google Scholar]
- Optically encoded multifunctional nanospheres for one-pot separation and detection of multiplex dna sequences. Anal. Chem.. 2013;85(24):11929-11935.
- [Google Scholar]
- Multiplexed fluorescence detection of microRNAs based on novel distinguishable quantum dot signal probes by cycle amplification strategy. Sens. Actuators B. 2017;252:1026-1034.
- [Google Scholar]
- Gold nanoparticle-based homogeneous fluorescent aptasensor for multiplex detection. Analyst. 2011;136(18):3720-3724.
- [Google Scholar]
- Krugman, S., Giles, J.P., Hammond, J.J.R.i.M.V. 1997. Infectious Hepatitis: Evidence for Two Distinctive Clinical, Epidemiological, and Immunological Types of Infection.Medical Virology, 7(3), 3-12
- Fluorescent “on-off-on” switching sensor based on CdTe quantum dots coupled with multiwalled carbon nanotubes@graphene oxide nanoribbons for simultaneous monitoring of dual foreign DNAs in transgenic soybean. Biosens. Bioelectron.. 2017;92:26-32.
- [Google Scholar]
- Simultaneous detection of three zoonotic pathogens based on phage display peptide and multicolor quantum dots. Anal. Biochem.. 2020;608:113854
- [Google Scholar]
- DNA adsorbed on graphene and graphene oxide: fundamental interactions, desorption and applications. Curr. Opin. Colloid Interface Sci.. 2016;26:41-49.
- [Google Scholar]
- Multiplex electrochemiluminescence DNA sensor for determination of hepatitis B virus and hepatitis C virus based on multicolor quantum dots and Au nanoparticles. Anal. Chim. Acta. 2016;916:92-101.
- [Google Scholar]
- Simultaneous visual detection of HAV and HBV based on hybrid molecular imprinting fluorescence sensor. Anal. Chem.. 2019;91:15748-15756.
- [Google Scholar]
- Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron.. 2018;100:89-95.
- [Google Scholar]
- Colorimetric detection of hepatitis B virus (HBV) DNA based on DNA-templated copper nanoclusters. Anal. Chim. Acta. 2016;909:101-108.
- [Google Scholar]
- A homogeneous and “off–on” fluorescence aptamer-based assay for chloramphenicol using vesicle quantum dot-gold colloid composite probes. Anal. Chim. Acta. 2016;929:49-55.
- [Google Scholar]
- Desorption of single-stranded nucleic acids from graphene oxide by disruption of hydrogen bonding. Analyst. 2013;138(6):1745-1749.
- [Google Scholar]
- A signal on-off fluorescence sensor based on the self-assembly DNA tetrahedron for simultaneous detection of ochratoxin A and aflatoxin B1. Anal. Chim. Acta. 2022;1198:339566
- [Google Scholar]
- Semaine, W., Johar, M., Tyrrell, D., Kumar, R., Agrawal, B.J.J.o.M.C. 2006. Inhibition of Hepatitis B Virus (HBV) Replication by Pyrimidines Bearing an Acyclic Moiety: Effect on Wild-Type and Mutant HBV. Journal of Medicinal Chemistry, 49(6), 2049-2054
- Recent advances in nanotechnology for simultaneous detection of multiple pathogenic bacteria. Nano Today. 2021;38:101121
- [Google Scholar]
- Exotic viral hepatitis; a review on epidemiology, pathogenesis, and treatment. J. Hepatol. 2022
- [CrossRef] [Google Scholar]
- Electrochemical detection of hepatitis B and papilloma virus DNAs using SWCNT array coated with gold nanoparticles. Biosens. Bioelectron.. 2013;41:205-210.
- [Google Scholar]
- Development and application of a universal Taqman real-time PCR for quantitation of duck hepatitis B virus DNA. J. Virol. Methods. 2013;191(1):41-47.
- [Google Scholar]
- Fabrication of fluorescent biosensing platform based on graphene oxide-DNA and their application in biomolecule detection. TrAC Trends Anal. Chem.. 2018;106:53-61.
- [Google Scholar]
- High-throughput SNP detection by using DNA pooling and denaturing high performance liquid chromatography (DHPLC) Hum. Genet.. 2000;107(5):483-487.
- [Google Scholar]
- Adsorption and desorption of DNA on graphene oxide studied by fluorescently labeled oligonucleotides. Langmuir. 2011;27(6):2731-2738.
- [Google Scholar]
- Single-excited double-emission CdTe@CdS quantum dots for use in a fluorometric hybridization assay for multiple tumor-related microRNAs. Microchim. Acta. 2020;187(2):134.
- [Google Scholar]
- An aptamer-mediated CdSe/ZnS QDs@graphene oxid composite fluorescent probe for specific detection of insulin. Sens. Actuators B. 2018;255:2339-2346.
- [Google Scholar]
- Highly sensitive detection of hepatitis C virus DNA by using a one-donor-four-acceptors FRET probe. Talanta. 2018;185:118-122.
- [Google Scholar]
- Fluorescence immunoassay for multiplex detection of organophosphate pesticides in agro-products based on signal amplification of gold nanoparticles and oligonucleotides. Food Chem.. 2020;326:126813
- [Google Scholar]
- A versatile graphene-based fluorescence “on/off” switch for multiplex detection of various targets. Biosens. Bioelectron.. 2011;26(7):3260-3265.
- [Google Scholar]
- An upconversion fluorescent resonant energy transfer biosensor for hepatitis B virus (HBV) DNA hybridization detection. Analyst. 2015;140(22):7622-7628.
- [Google Scholar]
- Label-free surface-enhanced Raman spectroscopy detection for tyrosine-methionine-aspartate-aspartate (YMDD)-motif mutants of HBV DNA. Vib. Spectrosc.. 2021;114:103253
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.104582.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







