Translate this page into:
Structural elucidation, antioxidant and hepatoprotective activities of chemical composition from Jinsi Huangju (Chrysanthemum morifolium) flowers
⁎Corresponding authors at: TCM and Ethnomedicine Innovation & Development International Laboratory, Innovative Material Medical Research Institute, School of Pharmacy, Hunan University of Chinese Medicine, Changsha 410208, China (W. Wang). cpujyq2010@163.com (Yu-Qing Jian), wangwei402@hotmail.com (Wei Wang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
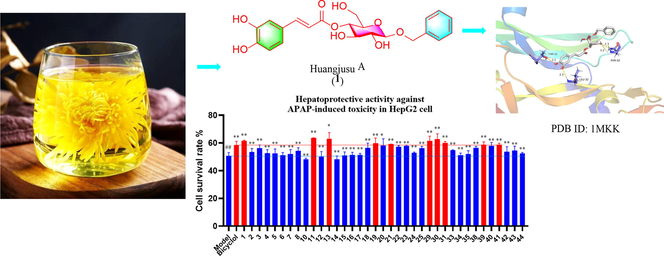
Abstract
Six new compounds (huangjusus A-F), including three caffeic acid glycosides (1–3), one quinic acid derivative (4), one dihydroflavone glycoside (11) and one monoterpene (31), together with thirty-eight known compounds (5–10, 12–30, 32–44), were obtained from “Jinsi Huangju” (Chrysanthemum morifolium Ramat.) flowers. Their structures were elucidated on the basis of the data obtained from different spectroscopic techniques. Among these compounds, five new (1–4 and 11) and ten known (5–9, 12, 13, 17, 40, and 42) compounds demonstrated significant 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) radical scavenging effects with IC50 values of 4.22–19.90 μM. Furthermore, three new compounds (1, 11, and 31) and seven known compounds (13, 19, 21, 29, 30, 39, and 41) exhibited potent hepatoprotective activities against N-acetyl-p-aminophenol (APAP)-induced toxicity in HepG2 cells with the cell survival rates of 61.53 %, 63.55 %, 60.01 %, 63.05 %, 59.75 %, 59.15 %, 61.07 %, 62.72 %, 58.86 %, and 58.76 % (positive control bicyclol, 58.41 %), respectively, at a concentration of 10 μM. These results indicate the potency of the flowers against radicals and in hepatoprotections.
Keywords
Jinsi Huangju (JH)
Chrysanthemum morifolium
Chemical constituents
Antioxidant activity
Hepatoprotective activity
Huangjusu
1 Introduction
“Jinsi Huangju” (Chrysanthemum morifolium Ramat.) belongs to the Compositae family and gets the name from its petal which looks like the gold spun (Jinsi in Chinese). It is a popular beverage, used as a health tea in China. Chrysanthemum is an important medicinal and edible plant, recognized worldwide (Chen et al., 2020). Traditional Chinese Medicine (TCM) practitioners have been utilizing this plant for thousands of years to treat digestive problems, fever, liver, and problems associated with eyes (Lin and Harnly, 2010; Ma et al., 2020). The increased use of chrysanthemums has subsequently led to its increased breeding and cultivation that intends to improve content of active compositions in chrysanthemums. During its long history of breeding, there are many classic and precious species of C. morifolium collectively termed chrysanthemum (Chen et al., 2020). “Jinsi Huangju” (JH) is one of valuable chrysanthemum because of its large flowers, extending up to 6 cm. One flower is enough to make a cup of tea.
The reported literature about phytochemicals and biological activities of JH is limited (Zhu et al., 2018; Liu et al., 2020). Previous investigations have led to the isolation of metabolites such as flavonoids, terpenoids, and phenylpropanoids from chrysanthemum (Yang et al., 2019; Yuan et al., 2020). Moreover, the extracts and compounds of the plants have exhibited antioxidant, anti-inflammatory, hepatoprotective, anti-microbial, neuroprotective, anti-tumor, and antiviral activities (Yang et al., 2017; Jiang et al., 2021). Most recently, high performance liquid chromatography (HPLC), ultra performance liquid chromatography/quadrupole time of flight mass spectrometry (UPLC‑QTOF‑MS/MS) and liquid chromatography mass spectrometry (LC/MS) techniques have been employed for the qualitative and quantitative analyses of these plants (Chen et al., 2020; Ma et al., 2020). However, these techniques may lead to inaccurate results due to the presence of structural isomers or enantiomers.
The aim of this work is to provide more information about the chemical constituents and biological potential of the flowers of JH. Hence, six new and thirty-eight known secondary metabolites (25 compounds were obtained for the first time) were isolated from this plant. Meanwhile, the results of the biological activities indicated that the isolated compounds possess antioxidant, hepatoprotective, and antibacterial effects. This article presents the isolation, identification, biological evaluation, and molecular docking studies of the secondary metabolites from JH flowers.
2 Materials and methods
2.1 General experimental procedures
The high-resolution-mass-spectrometry (HR-ESI-MS) data were acquired using a UPLC/xevo G2 QToF spectrometer (Waters Corporation, Milford, MA, USA). The optical rotations were measured on a Perking-Elmer 341-MC digital polarimeter at room temperature (PerkinElmer, Waltham, MA, USA). The infrared (IR) data were acquired utilizing a Hitachi 260–30 spectrometer (Hitachi High-Technologies Co., Japan). The electronic circular dichroism (ECD) data were determined on a Jasco J-810 Circular Dichroism spectropolarimeter at room temperature (Jasco, Mary's Court Easton, MD, USA). The nuclear magnetic resonance (NMR) data were obtained on a Bruker ARX-600 spectrometer (Bruker Technology Co., Ltd., Karlsruhe, Germany), using solvent peaks as references. Compounds were purified on an Agilent 1100 liquid chromatography (Agilent Technologies, Santa Clara, CA, USA) semi-preparative HPLC, equipped with an Agilent 1100 liquid chromatography (Agilent Technologies, Santa Clara, CA, USA) with an Agilent C18 (10 mm × 250 mm) column. Silica gel (80–100 mesh and 300–400 mesh) was got from Qingdao Haiyang Chemical Co. (Shandong, China). ABTS was purchased from Beijing Solarbio Science and Technology Co., Ltd (Beijing, China). HepG2 human liver cancer cell was got from American Type Culture Collection (ATCC) (USA). Three microbial strains, Staphylococcus aureus (ATCC 6538), Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 27853) were got from ATCC (USA).
2.2 Plant material
The flowers (9.9 kg) of “Jinsi Huangju” (Chrysanthemum morifolium Ramat.) were collected from Ningyuan county, Hunan province, China in November 2018. The plant was identified by Prof. Wei Wang and the voucher sample (the flowers, no. JSHJ2018-11) was deposited in the School of Pharmacy, Hunan University of Chinese Medicine.
2.3 Extraction and isolation
Dried flowers of “Jinsi Huangju” (9.9 kg) were immersed in 95 % ethanol (2 × 50L) at room temperature (each time 5 days) to give a crude extract. The ethanolic extract was suspended in water and successively partitioned with petroleum ether (PE), ethyl acetate (EA) and n-butanol. The PE layer (693.76 g) was subjected to silica gel column chromatography (CC) and eluted with PE-EA (100/1 to 0/1) to get seven fractions (A-G). Fraction F (134.04 g) was separated by a silica gel CC was eluted with a gradient PE/EA solvent system (100:1 to 0:1) to afford seven sub-fractions (PF1-PF7). PF2 (2.13 g) was further chromatographed on a silica gel column (PE-EA, 30:1 to 0:1) to get compound 16 (5.2 mg).
The EA extract (413.03 g) was then subjected to silica gel CC and eluted with CH2Cl2-MeOH (50:1 to 0:1) solvent system to get eleven fractions (EF1-EF11). Fraction EF5 (9.82 g) was separated by a YMC column, eluted successively with MeOH/H2O (60:40 to 100:0) to obtain ten sub-fractions (SF5.1–5.10). SF5.3 (5.2 g) was re-subjected to a YMC column with MeOH-H2O (10:90 to 100:0) to get further nine subfractions (5.3.1–5.3.9). SF5.3.4 (1.29 g) was then separated by silica gel column using gradient of PE/EA (10:1 → 0:1) and further purified by semi-preparative HPLC (flow rate: 3.0 ml/min) to obtain 33 (3.0 mg, tR = 13.2 min; CH3CN/H2O, 38:62), 34 (9.3 mg, tR = 20.1 min; CH3CN/H2O, 37:63), 35 (1.9 mg, tR = 15.0 min; CH3CN/H2O, 34:66), 28 (2.0 mg, tR = 16.2 min; CH3CN/H2O, 34:66), 24 (13.6 mg, tR = 14.1 min; CH3CN/H2O, 31:69), 20 (13.6 mg, tR = 15.2 min; CH3CN/H2O, 31:69), 22 (90.8 mg, tR = 17.2 min; CH3CN/H2O, 31:69), 21 (20.4 mg, tR = 20.4 min; CH3CN/H2O, 31:69), 23 (7.0 mg, tR = 15.6 min; CH3CN/H2O, 30:70), 38 (2.5 mg, tR = 17.5 min; CH3CN/H2O, 30:70), and 25 (3.1 mg, tR = 21.2 min; CH3CN/H2O, 25:75). SF5.4 (1.80 g) was loaded on a YMC column (MeOH-H2O, 50:50 to 100:0) to obtain six sub-fractions (SF5.4.1–5.4.6). SF5.4.3 (160.3 mg) was loaded on Sephadex LH-20CC (CH2Cl2/MeOH, 1:1) and further purified by semi-preparative HPLC (2.0 ml/min; CH3CN/H2O, 58:42) to give compounds 19 (1.0 mg, tR = 13.2 min), 17 (7.1 mg, tR = 17.5 min), 18 (1.5 mg, tR = 18.4 min), and 22 (1.7 mg, tR = 13.1 min; CH3CN/H2O, 55:45; 2.0 ml/min). EF7 (30.46 g) was chromatographed on YMC column (MeOH-H2O, 0:100 to 100:0) to afford eight sub-fractions (EF7.1–7.8). EF7.2 (2.90 g) was applied on a Sephadex LH-20 column (100 % MeOH) and further separated by semi-preparative HPLC (3.0 ml/min; CH3CN/H2O, 20:80) to get 42 (4.4 mg, tR = 7.7 min) and 43 (4.1 mg, tR = 10.8 min).
Fraction EF7.3 (8.15 g) was subjected to silica gel CC (PE/EA, 5:1 → 0:1) and then further purified by a semipreparative HPLC (3.0 ml/min) to obtain compounds 13 (5.1 mg, tR = 7.9 min; CH3CN/H2O, 32:68), 31 (4.2 mg, tR = 9.3 min; CH3CN/H2O, 30:70), 32 (4.1 mg, tR = 9.9 min; CH3CN/H2O, 28:72), 14 (2.1 mg, tR = 13.9 min; CH3CN/H2O, 21:79), 39 (28.9 mg, tR = 15.0 min; CH3CN/H2O, 21:79), 40 (12.1 mg, tR = 16.0 min; CH3CN/H2O, 21:79), 41 (3.0 mg, tR = 18.2 min; CH3CN/H2O, 21:79), 37 (7.0 mg, tR = 22.7 min; CH3CN/H2O, 21:79), 36 (4.0 mg, tR = 22.7 min; CH3CN/H2O, 21:79), 29 (11.7 mg, tR = 13.6 min; CH3CN/H2O, 20:80), and 30 (20.4 mg, tR = 13.5 min; CH3CN/H2O, 18:82).
Fraction EF7.4 (3.65 g) was separated by silica gel CC (PE/EA, 5:1 → 0:1) and then subjectedto a semi-preparative HPLC (3.0 ml/min) to obtain 1 (8.3 mg, tR = 15.1 min; CH3CN/H2O, 32:68), 8 (10.4 mg, tR = 21.2 min; CH3CN/H2O, 29:71), 7 (20.8 mg, tR = 26.4 min; CH3CN/H2O, 30:70), 44 (10.0 mg, tR = 26.4 min; CH3CN/H2O, 30:70), and 6 (20.8 mg, tR = 27.8 min; CH3CN/H2O, 30:70).
Fraction 7.5 (1.73 g) was subjected on a silica gel CC (PE/EA, 5:1 → 0:1) to afford compound 12 (4.5 mg) and sub-fractions 7.5.1 to 7.5.7. Fr. 7.5.3 (430.4 mg) was separated by Sephadex LH-20 column (100 % MeOH) and then processed on semi-preparative HPLC (3.0 ml/min) to obtain 2 (5.1 mg, tR = 22.3 min; CH3CN/H2O, 28:72), 3 (3.1 mg, tR = 24.1 min; CH3CN/H2O, 28:72), 4 (3.0 mg, tR = 30.6 min; CH3CN/H2O, 30:70), 5 (119.7 mg, tR = 22.6 min; CH3CN/H2O, 32:68), 9 (4.8 mg, tR = 23.1 min; CH3CN/H2O, 29:71), 10 (10.4 mg, tR = 28.7 min; CH3CN/H2O, 32:68), 11 (3.1 mg, tR = 22.4 min; CH3CN/H2O, 30:70), 26 (20.1 mg, tR = 18.1 min; CH3CN/H2O, 28:72), 27 (18.6 mg, tR = 18.1 min; CH3CN/H2O, 28:72). Fraction 11 (28.32 g) was separated on a YMC column (MeOH/H2O, 0:100 → 100:0) to give compound 15 (27.5 mg).
2.3.1 Huangjusu A (1)
Light brown solid.
− 38.0 (c 0.10, CH3OH); IR νmax: 3360, 1676, 1599, 1261, 1155, 1024, 699 cm−1; HR-ESI-MS (m/z 455.1314 [M + Na]+ and 887.2770 [2 M + Na]+, calcd for C22H24O9Na+, 455.1318); 1H NMR (600 MHz) and 13C NMR (150 MHz) data in CD3OD, see Tables 1 and 2.
1
2
3
4
11
31
NO.
δH (J in Hz)
1
3.90 m; 3.58 m
2.11 s
2
7.44 d (7.8)
7.27 m
2.40 m
2.30 dd (13.2, 3.6)
2.13 dd (13.8, 7.8)5.34 m
3
7.34 t (7.8)
7.27 m
5.40 dt (10.8, 1.8)
5.34 m
3.13 d (16.8, 12.6)
2.76 d (16.8, 8.0)2.57 m
4
7.28 t (7.8)
7.18 m
5.48 dt (10.8, 1.8)
3.93 m
2.36 m
5
7.34 t (7.8)
7.27 m
2.08 m
5.31 m
6
7.44 d (7.8)
7.27 m
0.98 t (7.8)
2.33 m; 2.17 m
6.20 s
7
4.96 d (12.0)
4.70 d (12.0)2.96 m
2.95 d (7.2)
8
4.12 m
3.79 m
6.23 s
5.55 m
9
5.46 m
10
3.25 d (7.2)
11
1′
4.43 d (7.8)
4.38 d (7.8)
4.35 d (7.8)
3.68 s
2′
3.37 m
3.31 m
3.28 m
7.44 d (1.8)
6.79 m
3′
3.62 m
3.64 m
3.62 m
4′
4.86 m
4.85 m
4.86 m
5′
3.51 m
3.52 m
3.52 m
6.74 d (8.4)
6.79 m
6′
3.64 m
3.58 m3.63 m
3.55 m3.62 m
3.55 m6.97 dd (8.4, 1.8)
6.92 m
7′
6.81 d (12.6)
8′
6.72 d (12.6)
9′
1′'
5.04 m
2′'
7.05 d (1.8)
7.05 d, (2.4)
7.05, d (1.8)
7.07 d (1.8)
3.53 m
3′'
3.69 m
4′'
4.89 m
5′'
6.78 d (8.4)
6.78 d (7.8)
6.78 d (8.4)
6.78 d (8.4)
3.69 m
6′'
6.96 dd (8.4, 1.8)
6.96 d (7.8, 2.4)
6.96 dd (8.4, 1.8)
6.97 dd (8.4, 1.8)
3.61 m; 3.51 m
7′'
7.60 d (15.6)
7.59 d (16.2)
7.60 d (15.6)
7.62 d (15.6)
8′'
6.30 d (15.6)
6.30 d (16.2)
6.30 d (15.6)
6.34 d (15.6)
9′'
1′''
4.20 m; 4.12 m
2′''
1.25 t (7.2)
5.92 d (15.6)
3′''
7.06 m
4′''
1.91 d (6.6)
1
2
3
4
11
31
NO.
δC
1
139.0
140.0
70.6
74.7
17.4
2
129.2
129.4
28.8
35.7
80.8
174.8
3
129.3
130.0
125.8
72.2
44.2
32.6
4
128.7
127.2
134.6
70.2
198.6
35.2
5
129.3
130.0
21.5
71.9
165.0
211.9
6
129.2
129.4
14.6
35.6
98.0
139.3
7
71.8
37.2
175.3
166.9
22.1
8
71.8
96.9
123.1
9
164.6
130.2
10
105.0
33.4
11
174.0
1′
103.3
104.4
104.4
127.9
131.5
52.3
2′
75.3
75.2
75.2
118.7
119.4
3′
75.8
75.8
75.9
145.7
146.6
4′
72.6
72.5
72.5
148.5
147.0
5′
76.2
76.2
76.1
115.7
116.2
6′
62.6
62.5
62.5
125.3
114.8
7′
145.8
8′
116.2
9′
167.1
1′'
127.7
127.7
127.7
128.1
101.1
2′'
115.2
115.2
115.2
115.1
74.8
3′'
146.9
146.9
146.9
146.8
75.5
4′'
149.7
149.7
149.7
149.5
71.9
5′'
116.5
116.5
116.5
116.5
76.3
6′'
123.1
123.1
123.1
123.0
62.0
7′'
147.6
147.6
147.6
147.1
8′'
114.7
114.7
114.7
115.1
9′'
168.6
168.6
168.6
168.8
1′''
62.6
167.3
2′''
14.4
123.1
3′''
147.4
4′''
18.1
2.3.2 Huangjusu B (2)
Light brown solid. − 29.0 (c 0.11, CH3OH); IR νmax: 3363, 1680, 1601, 1261, 1156, 1027, 701 cm−1; HR-ESI-MS (m/z: 469.1481 [M + Na]+ and 915.3050 [2 M + Na]+, calcd. for C23H26O9Na, 469.1475); 1H NMR (600 MHz) and 13C NMR (150 MHz) data in CD3OD, see Tables 1 and 2.
2.3.3 Huangjusu C (3)
Light brown solid. − 42.1 (c 0.10, CH3OH); IR νmax: 3410, 2964, 1700, 1603, 1262, 1162, 1080 cm−1; HR-ESI-MS (m/z: 447.1636 [M + Na]+, calcd. for C21H28O9Na, 447.1631); 1H NMR (600 MHz) and 13C NMR (150 MHz) data in CD3OD, see Tables 1 and 2.
2.3.4 Huangjusu D (4)
Light brown solid. − 83.4 (c 0.42, CH3OH); IR νmax: 3388, 1686, 1601, 1516, 1259, 1179, 1159, 1023 cm−1; HR-ESI-MS (545.1660 [M + H]+, calcd. for C27H29O12+, 545.1659); 1H NMR (600 MHz) and 13C NMR (150 MHz) data in CD3OD, see Tables 1 and 2.
2.3.5 Huangjusu E (11)
Light yellow solid. − 42.0 (c 0.10, CH3OH); IR νmax: 3423, 2360, 1684, 1205, 1138, 726, 634 cm−1; ECD (0.10 mg/ml, CH3OH) λmax (Δε) 294 (−3.06), 331 (+0.12); HR-ESI-MS data (m/z 541.1328 [M + Na]+ and 1059.2746 [2 M + Na]+, calcd. for C25H26O12Na+, 541.1322); 1H NMR (600 MHz) and 13C NMR (150 MHz) data in CD3OD, see Tables 1 and 2.
2.3.6 Huangjusu F (31)
Colorless oil. IR νmax: 2948, 1736, 1696, 1595, 1436, 1270, 1169 cm−1; (+)-HR-ESI-MS data (m/z 231.0989 [M + Na]+, calcd. for C12H16O3Na+, 231.0997); 1H NMR (600 MHz) and 13C NMR (150 MHz) data in CD3OD, see Tables 1 and 2.
2.4 ABTS radical scavenging assay
Antioxidant capacity of the samples was evaluated through the ABTS assay was evaluated as previously published literature (Yang et al., 2016). The ABTS radical scavenging ability (RSA) of the compounds was determined as follows:
l-Ascorbic acid (VC) was used as a reference standard. All results show the average values of experiments performed in triplicate.
2.5 Hepatoprotective activity
HepG2 cells were used to evaluate the hepatoprotective capacity of the compounds of JH, employing the method performed as described previously (Hao et al., 2012). The cells were cultured in 96-well plates (1 × 105 cells/well) overnight. 10 μM test samples, 10 μM bicyclol (positive control), and 6 mM APAP were added to the wells and incubated for another 48 h. The cell effect was evaluated with the help of 3-(4,5)-dimethylthiahiazo(-z-y1)-3,5-diphenytetrazoliumromide (MTT) assay (Hao et al., 2012). The optical density (OD) of the formazan solution was assessed using a microplate reader at 490 nm. Bicyclol was used as a reference standard.
2.6 Antibacterial activity evaluation
The extracts and compounds were tested against one gram-positive bacteria (S. aureus ATCC 6538) and two gram-negative bacteria (E. coli ATCC 25,922 and P. aeruginosa ATCC 27853) by a twofold serial dilution method in a 96-well microtiter plate, as reported previously (Jiang et al., 2019; Wei et al., 2019). Oxacillin was used as a reference standard. The minimum inhibitory concentration (MIC) values were calculated.
2.7 Network pharmacology analysis
2.7.1 Screening of potential targets of JH-1 ∼ 9 and non-alcoholic liver injury
TCMSP (https://tcmspw.com/tcmsp.php) database screening parameters of oral bioavailability (OB) ≥ 30 % and drug-likeness (DL) ≥ 0.18 resulted in the discovery of nine active compounds isolated from JH, including (+)-eriodictyol (JH-1), (2R,3R)-taxifolin (JH-2), apigenin (JH-3), apigenin-7-O-β-d-glucopyranoside (JH-4), artemetin (JH-5), jaceosidin (JH-6), geniposide (JH-7), protocatechuic acid (JH-8), p-hydroxybenzoic acid (JH-9). The genes associated with non-alcoholic liver injury were retrieved from Disgenet database (https://www.disgenet.org/), Drugbank database (https://www.drugbank.ca/) and String database (https://string-db.org/) (Gong et al., 2021). These genes were standardized by using Uniprot protein database (https://www.uniprot.org/) to get their official names. The common targets of non-alcoholic liver injury and JH-1 ∼ 9 were obtained using the online Venn diagram (https://bioinformatics.psb.ugent.be/webtools/Venn/).
2.7.2 Gene ontology (Go) and kyoto encyclopedia of genes and genomes (KEGG) analysis
The key targets of non-alcoholic liver injury and JH-1 ∼ 9 were imported into Metascape (https://metascape.org/gp/index.html#/main/step1) for gene functional annotation. GO and KEGG analyses were performed on the obtained results by selecting GO Biological Processes, GO Molecular Functions, GO Cellular Components, and KEGG Pathway. The top 20 items were selected (P < 0.05), and R statistical language (Version: 3.3.2) was applied to plot the GO and KEGG enrichment pathways.
2.7.3 Development of protein–protein interaction network
The selected common targets were built by protein–protein interaction (PPI) network using the STRING database (https://string-db.org/). Subsequently, we imported the targets information into Cytoscape software (Version: 3.6.1) to further construct the core PPI network by MCC, DMNC, MNC, Degree, EPC, BottleNeck, EcCenTricity, Closeness, Radiality, Betweenness, Stress and ClusteringCoefficient analysis. The network analyzer function in the CytoNCA plug-in was then used to investigate the topological properties of the PPI network. Lastly, the PPI network diagram was founded.
2.8 Molecular docking study
The crystal structure of vascular endothelial growth factor A (VEGFA) (PDB ID: 1MKK) was downloaded from the RCSB Protein Data Bank (Gong et al., 2021). The 3D structures of compounds 1 and 13 were drawn by Chem3D program. Docking was done using AutoDockTools (version 1.5.6) and structure visualization was analyzed by Pymol (v. 2.3.2) and Discovery Studio client 2020.
2.9 Statistical analysis
The tests were performed in triplicate, and the results shown as mean ± standard deviation (SD). The data was calculated using Graph prism 8.0 software with analysis of variance; at the same time, p < 0.05 and p < 0.01 was identified statistically significant.
3 Results and discussion
3.1 Structure elucidation
Chromatographic separation of the PE and EtOAc layers of JH flowers resulted in the isolation of six new compounds (1–4, 11 and 31) along with thirty-eight known compounds (5–10, 12–30 and 32–44) (Fig. 1).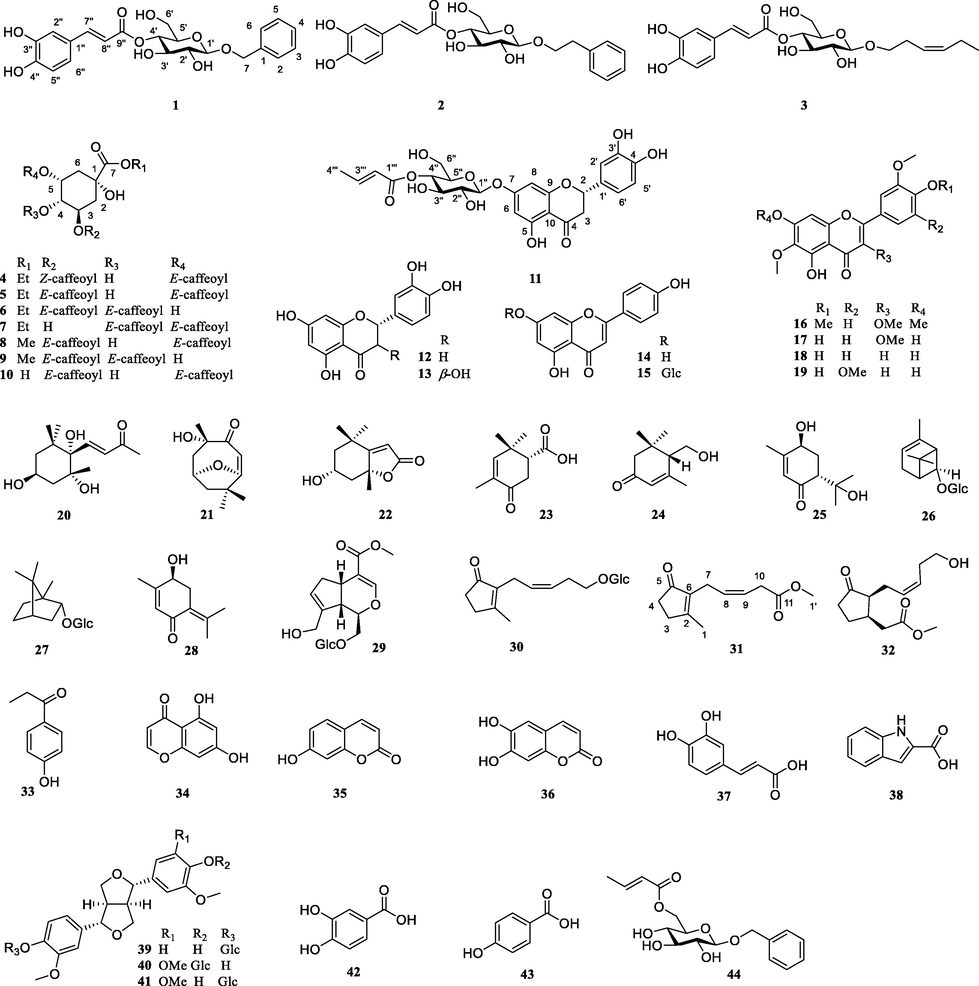
The structures of compounds 1–44.
Huangjusu A (1), was yielded as a light brown solid. Its molecular formula was established to be C22H24O9 from the (+)-HR-ESI-MS analysis, showing a signal at m/z 455.1314 [M + Na]+ and 887.2770 [2 M + Na]+ (calcd for C22H24O9Na+, 455.1318), indicating 11 degrees of unsaturation. The 1H NMR spectra of 1 displayed the presence of a benzyl moiety [δH 7.44 (2H, d, J = 7.8 Hz), 7.34 (2H, t, J = 7.8 Hz),7.28 (1H, t, J = 7.8 Hz) and δH 4.96 (1H, d, J = 12.0 Hz), 4.70 (1H, d, J = 12.0 Hz)] and an E-caffeoyl moiety [δH 7.05 (1H, d, J = 1.8 Hz), 6.96 (1H, dd, J = 8.4, 1.8 Hz), 6.78.
(1H, d, J = 8.4 Hz), δH 7.60 (1H, d, J = 15.6 Hz) and 6.30 (1H, d, J = 15.6 Hz)]. Moreover, the 1H NMR data of 1 displayed resonance signals for a glucosyl moiety [δH 4.86 (1H, overlapped), 4.43 (1H, d, J = 7.8 Hz), 3.62 (1H, m), 3.51 (1H, m), 3.37 (1H, m) and 3.64 (1H, m), 3.58 (1H, m)]. The 13C NMR spectra showed 22 carbon signals, including a carbonyl carbon (δc 168.6), 12 aromatic carbons (2 benzene rings), 2 olefinic carbons, 5 oxygenated methine carbons (δC 103.3, 76.2, 75.8, 75.3 and 72.6), and 2 oxygenated methylene carbons (δC 71.8 and 62.6). The 1D NMR spectrum (Table 1 and 2) inferred that the structure of 1 was similar to 1-O-benzyl-6-O-E- caffeoyl-β-d-glucopyranoside (Janggyoo et al., 2015), except for the E-caffeoyl group in place of glucosyl group. The result was confirmed by the 1H–1H COSY connectivity of H-1′ (δH 4.86)/H-2′ (δH 3.37)/H-3′ (δH 3.62)/H-4′ (δH 4.86)/H-5′ (δH 3.51)/H-6′ (δH 3.64, 3.65) and the HMBC correlations from H-4′ and H-7′' (δH 7.60) to C-9′' (δC 168.6) (Fig. 2). Furthermore, the sugar group was identified as d-glucoside (tR = 17.19 min) by acid hydrolysis and HPLC analysis (Fig. S1). Hence, the structure of 1 was confirmed to be 1′-O-benzyl-4′-O-E-caffeoyl-β-d-glucopyranoside and named huangjusu A.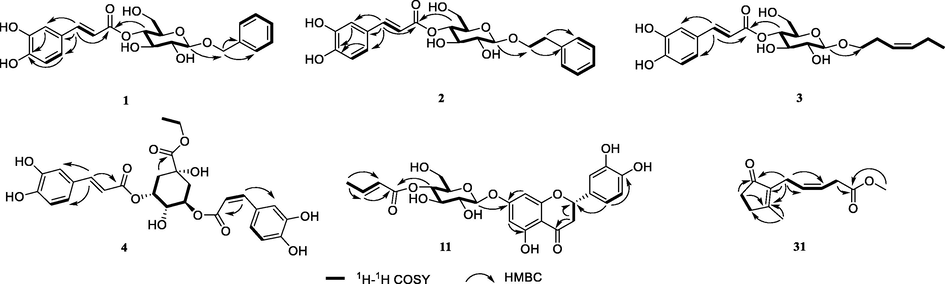
The key 1H−1H COSY and HMBC correlations of compounds 1–4, 11 and 31.
Compound 2, a light brown solid, possessed the molecular formula C23H26O9 according to its (+)-HR-ESI-MS (m/z: 469.1481 [M + Na]+ and 915.3050 [2 M + Na]+, calcd. for C23H26O9Na, 469.1475). The 1D NMR spectra of 2 showed high similarities to that of compound 1. An additional methylene group was elucidated to be present between C-1 and C-7. This was confirmed by the HMBC correlations of H-7 (δH 2.96) with C-2,6 (δC 129.4), C-1 (δC 140.0) and C-8 (δC 71.8). The molecular formula of 2 further confirmed that it had an extra methylene moiety. Hence, the structure of 2 was confirmed to be 1′-O-phenethyl-4′-O-E-caffeoyl-β-d-glucopyranoside, named as huangjusu B.
Compound 3, a light yellow solid, possessed the molecular formula C21H28O9 according to its (+)-HR-ESI-MS (m/z: 447.1636 [M + Na]+, calcd. for C21H28O9Na, 447.1631). The 1D NMR spectra of 3 exhibited similarities to that of compound 1, except for the benzyl moiety replaced by cis-3-hexenyl moiety at C-1′. This was confirmed by the 1D NMR and 2D NMR data. The 1D NMR spectra displayed signals for the cis-3-hexenyl moiety [δH 3.90 (H-1a), δH 3.58 (H-1b)/δC 70.6 (C-1), δH 2.40 (H-2)/δC 28.8 (C-2), δH 5.40 (1H, dt, J = 10.8, 1.8 Hz, H-3)/δC 125.8 (C-3), δH 5.48 (1H, dt, J = 10.8, 1.8 Hz, H-4)/δC 134.6 (C-4), δH 2.08 (H-5)/δC 21.5 (C-5) and δH 0.98 (H-6)/δC 14.6 (C-6)]. Furthermore, this was further confirmed by the HMBC correlations of H-1′ (δH 4.35) with C-1 (δC 70.6). Hence, the structure of 3 was confirmed and named as huangjusu C.
Compound 4 was obtained as a light yellow solid. Its molecular formula was assigned to be C27H28O12 from its (+)-HR-ESI-MS data (m/z 545.1660 [M + H]+, calcd for C27H29O12+, 545.1659). The 1D NMR data of 4 was similar to ethyl-3,5-di-O-caffeoylquinate (5) (Wang et al., 1992), the only difference being the replacement of the trans-olefinic protons with a large coupling constant [δH 7.55 (1H, d, J = 15.9 Hz, H-7′), 6.23 (1H, d, J = 15.9 Hz, H-8′)] by cis-olefinic protons with a smaller coupling constant [δH 6.81 (1H, d, J = 12.6 Hz, H-7′), 6.72 (1H, d, J = 12.6 Hz, H-8′)]. Thus, the structure of 4 was confirmed and named as huangjusu D.
Compound 11 was purified as a pale-yellow powder. Its molecular formula was assigned to be C25H26O12 from its (+)-HR-ESI-MS data (m/z 541.1328 [M + Na]+ and 1059.2746 [2 M + Na]+, calcd for C25H26O12Na+, 541.1322), indicating 13 degrees of unsaturation. The 1H NMR spectra of 11 exhibited typical 5,7-substituted ring A resonances of flavonoids at δH 6.23 (1H, m), 6.20 (1H, m); 1′,3′,4′-substituted ring B resonances at δH 6.92 (1H, m), 6.89 (2H, m); an oxygenated methine [δH 5.34 (1H, m)], and a methylene [δH 3.13 (1H, d, J = 16.8, 12.6 Hz) and 2.76 (1H, d, J = 16.8, 8.0 Hz] of the ring C of flavonone. Furthermore, the 1H NMR data of 11 also illustrated a sugar moiety [δH 5.04 (1H, m), 4.89 (1H, m), 3.69 (1H, m), 3.69 (1H, m), 3.53 (1H, m), and 3.61 (1H, m), 3.51 (1H, m)], and a 2-E-butenoate moiety [δH 7.06 (1H, m), 5.92 (1H, d, J = 15.6 Hz) and 1.91 (1H, d, J = 6.6 Hz)]. According to the above data, compound 11 revealed existence of a flavonone unit, a glucopyranosyl unit and a 2-E-butenoate group. Their linkages were established from the 2D NMR data. HMBC experiment revealed that H-1′' (δH 5.04), H-8 (δH 6.23) and H-6 (δH 6.20) all have correlations to C-7 (δC 166.9). This confirmed the attachment of the β-glucose unit with flavonone at C-7. Other key HMBC relationships from H-4′' (δH 4.89), and H-3′'' (δH 7.06) to C-1′'' (δC 167.3) deduced that the 2-E-butenoate moiety would be attached to C-4′' of the β-glucose moiety. Moreover, on the basis of the negative cotton effect at 290 nm and positive cotton effect at 334 nm in the experimental ECD spectrum, the absolute configuration at C-2 of the flavonone aglycone was determined to be S (Slade et al., 2005). Therefore, the structure of 11 was ascertained as (+)-eriodictyol 7-O-(4′'-O-2-E-butenoate)-β-glucopyranoside and named huangjusu E.
Compound 31, a colorless oil, possessed the molecular formula C12H16O3 concluded by (+)-HR-ESI-MS (m/z: 231.0989 [M + Na]+, calcd for C12H16O3Na, 231.0997). The 1D NMR data of 31 was similar to cis-jasmone (Birkett et al., 2000), except for the methyl ester moiety at C-11 in 31 as compared to methyl group in cis-jasmone. This outcome was confirmed by the HMBC correlations of H-10 (δH 3.25) and H-1′ (δH 3.68) with C-11 (δC 174.0) and of H-10 with C-9 (δC 130.2) and C-8 (δC 123.1) and the molecular weight of the compound. Thus, the structure of 31 was ascertained and named huangjusu F.
In addition, the structures of 38 known compounds were determined as ethyl 3,5-di-O-caffeoylquinate (5) (Wang et al., 1992), ethyl 3,4-di-O-caffeoylquinate (6) (Wang et al., 1992), ethyl 4,5-di-O-caffeoylquinate (7) (Guan et al., 2014), methyl 3,5-di-O-caffeoylquinate (8) (Liu et al., 2010), methyl 3,4-di-O-caffeoylquinate (9) (Liu et al., 2010), 3,5-di-O-caffeoylqunic acid (10) (Wang et al., 1992), (+)-eriodictyol (12) (Huvaere et al., 2012), (2R,3R)-taxifolin (13) (Huvaere et al., 2012), apigenin (14) (Wei et al., 2019), apigenin-7-O-β-d-glucopyranoside (15) (Wei et al., 2019), artemetin (16) (Hussain et al., 2009), jaceidin (17) (Hussain et al., 2009), jaceosidin (18) (Martínez et al., 1987), 6-methoxytricin (19) (Martínez et al., 1987), (3S,5R,6S,7E)-3,5,6-trihydroxy-7-megastigmen-9-one (20) (Sun et al., 2007), pubinernoid A (21) (Wan et al., 2017), isololiolide (22) (Sun et al., 2007), (1R)-3-oxo-4,6,6-trimethylcyclohex-4-ene-l-carboxylic acid (23) (Marco et al., 1991), crocusatin C (24) (Wook et al., 2016), (+)-trans-sobrerol (25) (Pellegata et al., 1987), (-)-cis-chrysanthenol O-β-d-glucopyranoside (26) (Miyakado et al., 1974), (-)-borneol 2-O-β-d-glucopyranoside (27) (Orihara and Furuya, 1993), 4-hydroxypiperitenone (28) (Thulasiram and Madyastha, 2005), geniposide (29) (Ono et al., 2005), (Z)-5′-hydroxyjasmone 5′-O-β-d-glucopyranoside (30) (Ding et al., 2009), tuberonic acid (32) (He et al., 2019), 4-propionylphenol (33) (Ma et al., 2021), 5,7-dihydroxy-4H-4-chromenone (34) (Kwon et al., 2016), umbelliferone (35) (Ahn et al., 2021), aeculetin (36) (Ahn et al., 2021), caffeic acid (37) (Ahn et al., 2021), 1H-indole-2-carboxylic acid (38) (Silva et al., 2021), pinoresinol-4-O-glucoside (39) (Deyama et al., 1986), eucommin A (40) (Deyama et al., 1986), (+)-isoeucommin A (41) (Deyama et al., 1986), protocatechuic acid (42) (Ahn et al., 2021), p-hydroxybenzoic acid (43) (Ahn et al., 2021), benzyl-6-[(2E)-2-butenoate]-β-d-glucopyranoside (44) (Qi et al., 2011) by comparison of their 1D NMR data with those described in the literature (showed in Supplement data).
3.2 Antioxidant activity
Some of the isolated compounds were tested for their antioxidant potential by employing the ABTS radical cation scavenging assay according to the procedures as previously described. As shown in Table 3, the new compounds huangjusus A-E (1–4 and 11) exhibited strong scavenging effect with IC50 values of 11.34 μM, 15.24 μM and 10.32 μM, 6.38 μM, and 10.23 μM, respectively. Eleven known compounds (5–9, 12, 13, 17, 39, 40, and 42) showed moderate to potent antioxidant effects with IC50 values of 4.22–56.33 μM.
Compounds
ABTSa,b (μM)
ascorbic acid (Vc)
22.00 ± 1.29
1
11.34 ± 1.56*
2
15.24 ± 1.80*
3
10.32 ± 1.71*
4
6.38 ± 1.43*
5
8.39 ± 1.28*
6
4.22 ± 1.25*
7
5.95 ± 2.09*
8
6.43 ± 1.67*
9
7.78 ± 1.14*
11
10.23 ± 1.48*
12
4.70 ± 1.28*
13
8.65 ± 1.76*
17
9.13 ± 1.42*
39
56.33 ± 2.12*
40
19.90 ± 2.05*
42
12.61 ± 1.49*
43
103.02 ± 1.86*
Structure and antioxidant activity relationship study (SARs) showed that the introduction of two adjacent phenolic hydroxyl moieties could obviously enhance the activity (comparing the activity of 42 and 43). Neither the alcoholic hydroxyl group nor a single phenolic hydroxyl group showed potent activity.
3.3 Hepatoprotective activity
The compounds were tested for hepatoprotective activity using MTT assay. As shown in Fig. 3, most of these compounds (except 8, 10, and 14) improved the survival rate of APAP-triggered toxicity in HepG2 cells compared with the model group. Among them, ten compounds (1, 11, 13, 19, 21, 29, 30, 31, 39, and 41) exhibited potent hepatoprotective activities against APAP-induced toxicity in HepG2 cells with the cell survival rates of 61.53 %, 63.55 %, 63.05 %, 59.75 %, 59.15 %, 61.07 %, 62.72 %, 60.01 %, 58.86 %, and 58.76 % (positive control bicyclol, 58.41 %) at 10 μM, respectively.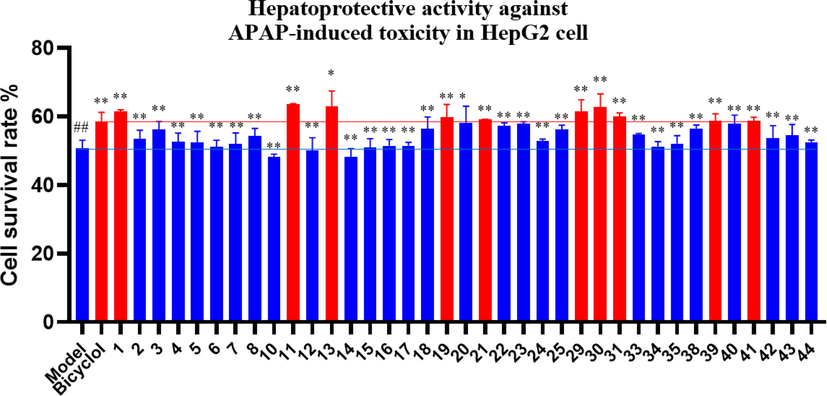
Activities of some compounds on the survival rates of APAP-induced HepG2 cells. Data are presented as the mean ± SD (n = 3). Values with the “##”, “**” and “*” are significantly different (## p < 0.01 vs blank; ** p < 0.01, * p < 0.05 vs model).
3.4 Network pharmacology analysis
3.4.1 Screening of the common targets
According to database analysis, 65 targets of JH-1 ∼ 9 and 1234 targets of non-alcoholic liver injury were obtained. Duplicate results were removed. The Venn diagram was produced using the online Venn analysis tool, and a total of 52 common targets were found to be the potential common targets of JH-1 ∼ 9 and non-alcoholic liver injury (Fig. 4A and 4B).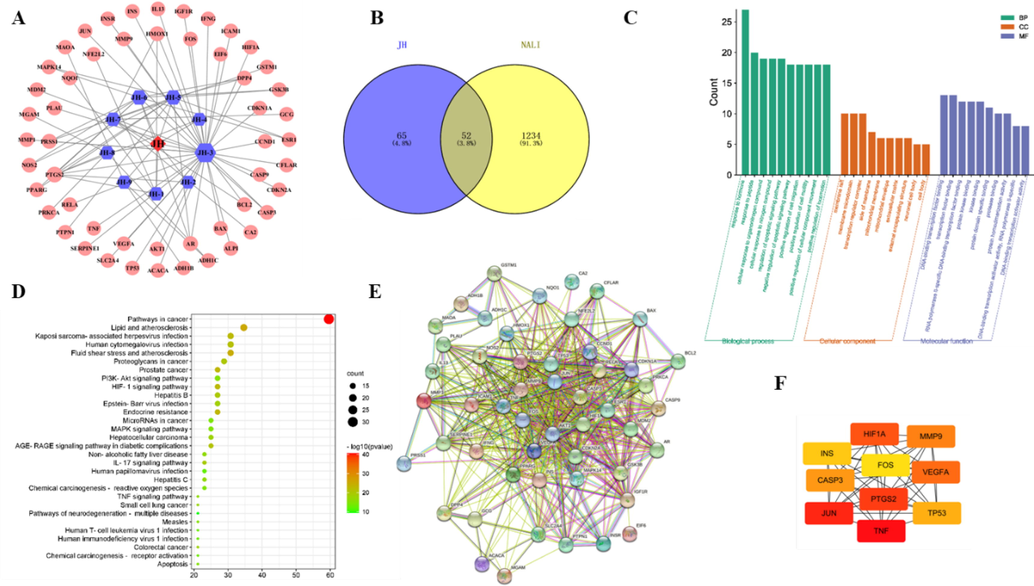
Network pharmacology analysis of JH-1 ∼ 9 and non-alcoholic liver injury. (A) Chrysanthemum-component-target network diagram. (B) Venn diagram of the com-mon targets of JH-1 ∼ 9 and non-alcoholic liver injury. (C) GO enrichment analysis of the predicted targets in Biological Process, Molecular Function and Cell Composition. (D) Bubble plot of the potential targets obtained from KEGG enrichment analysis. (E) Protein-protein target interaction network of the common targets. (F) The core target interaction diagram.
3.4.2 GO and KEGG analysis
The 52 key targets were imported into the online Metascape database (https://metascape.org/gp/index.html#/main/step1) which resulted in identification of 969 biological processes, 29 cell components, and 75 molecular functions by GO analysis. The top 20 items are shown in Fig. 4D. KEGG pathway enrichment results exhibited that there were 157 signaling pathways and the top 30 items were illustrated in Fig. 4C, which were closely related to proteoglycans in cancer, pathways in cancer, and PI3K/Akt signaling pathway (see Fig. 4).
3.4.3 Protein-protein interaction network construction
The 52 potential targets shared by JH-1 ∼ 9 and non-alcoholic liver injury were imported into String database to obtain the protein–protein interaction (PPI) network (Fig. 4E). PPI results showed that the network had 51 nodes and 562 edges. The PPI analysis results of the potential targets were saved as TSV format and imported into Cytoscape software for further analysis. According to the 12 kinds of algorithm, including MCC, DMNC, MNC, Degree, EPC, BottleNeck, EcCenTricity, Closeness, Radiality, Betweenness, Stress, ClusteringCoefficient analysis, the degree of 10 targets were greater than the given value, exhibiting the key nodes of JH-1 ∼ 9. These core targets might be the potential targets of JH-1 ∼ 9 in the treatment of liver injury (Fig. 4F).
3.5 Molecular docking
According to the results of hepatoprotective activity and the potential core targets obtained from network pharmacology analysis, molecular docking was utilized to predict the possible binding ability between the evaluated huangjusu A (1) and 13 and the VEGFA (Gong et al., 2021). The computing results of molecular docking are presented in Fig. 5. Huangjusu A (1) formed hydrogen-bonding interaction with Leu-32, Leu-66, Thr-31, and Asn-62 residues and hydrophobic interactions with Leu-32 and Ile-29 (Fig. 5A). Compound 13 showed hydrogen-bonding interactions with Asp-34, Ala-61, Phe-47, and Ile-43, and the hydrophobic interaction with Asp-34 and Ile-46 (Fig. 5B). These results indicated that the hydroxy and benzene ring play a major role in the docking, moreover, pharmacological mechanisms need to be further explored (Hung et al., 2022).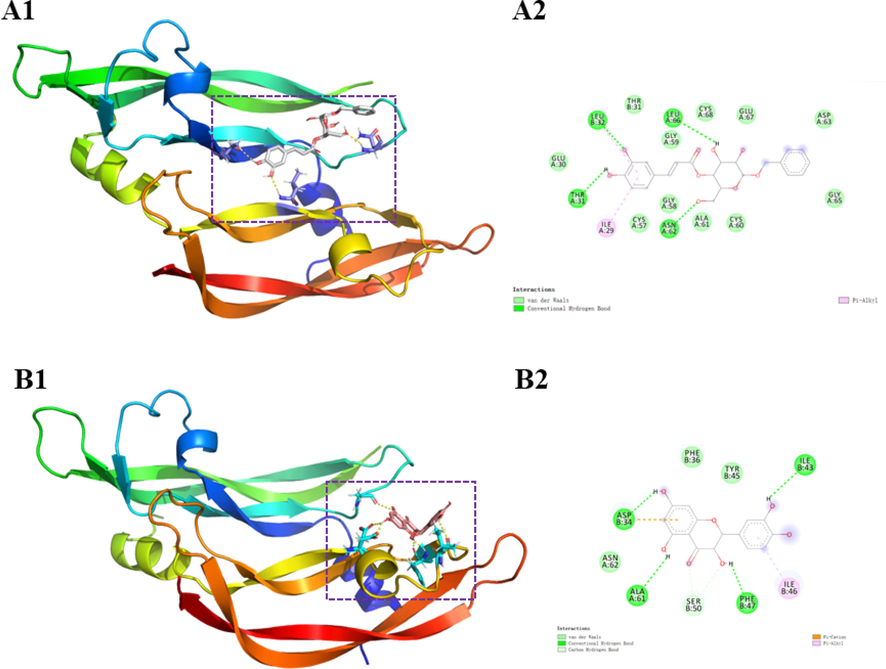
Docking picture of compounds 1 (A) and 13 (B).
3.6 Antibacterial activity
As an attempt to illustrate the antibacterial effect of compounds and crude extract of JH, a radial diffusion assay was firstly applied on a sensitive strain of S. aureus, E. coli, and P. aeruginosa. Compounds and extracts with anti-microbial activity were indicated by the inhibition zones observed on the agar plate.(Wei et al., 2019) To establish the MIC values, the samples were subjected to a series of liquid dilution techniques on a 96-well microtitration plate.
As shown in Supplementary Table S1, the crude extract of JH and some compounds exhibited moderate and weak anti-bacterial effects against all the selected strains. Compound 19 displayed significant inhibition of S. aureus with MIC values of 22.5 μg/mL. Moreover, ethanol and PE extracts exhibited weak activity with MIC values of 250.0 and 500 μg/mL, respectively. Compounds 5, 6, 8, 15, 13, 20, 21, and 35 showed selected activity against P. aeruginosa with MIC values of 34.0–121.0 μg/mL, compared to the positive control oxacillin (IC50 = 100.0 μg/mL). Among these, compounds 5, 6, and 7 are ethyl dicaffeoylquinates which have the same molecular weight, but their antibacterial activities were different. Ethyl 3,5-di-O-caffeoylquinate (5) and ethyl 3,4-di-O-caffeoylquinate (6) exhibited weak to moderate antibacterial activities, but ethyl 4,5-di-O-caffeoylquinate (7) showed no activity, indicating that the caffeoyl group at C-3 of quinine ethyl carbonate is critical for antibacterial activity. Additionally, both ethyl 3,5-di-O-caffeoylquinate (5) and methyl 3,5-di-O-caffeoylquinate (8) showed weak to moderate antibacterial activities, while 3,5-di-O-caffeoylqunic acid (10) had no activity, demonstrating that the carboxyl substituent of quinic acid also has a great influence on the antibacterial activity. No compounds or extracts showed strong inhibitory effects against the E. coli strain.
4 Conclusions
Phytochemical analysis, in search of bioactive metabolites from the flowers of JH, led to the isolation and characterization of six new and 38 known secondary metabolites. The isolated compounds were characterized as caffeic acid derivatives (caffeic acid glycoside and caffeoylquinic acid), flavonoids, monoterpenes, phenylpropanoids (simple phenylpropanoids and lignans) and others. Some of these compounds exhibited significant or moderate antioxidant, hepatoprotective, and antibacterial activities. The five new compounds huangjusus A-D (1–4 and 11) showed significant antioxidant activities owing to two adjacent phenolic hydroxyl groups through SARs. It was thus concluded that the caffeic acid derivatives (caffeic acid glycosides and caffeoylquinic acids) and some flavonoids of JH might be responsible for the antioxidant potential of the flowers. Furthermore, three new compounds (1, 11, and 31) and seven known compounds (13, 19, 21, 29, 30, 39, and 41) showed strong hepatoprotective effects. Among them, six glycosides (1, 11, 29, 30, 39, and 41) exhibited strong hepatoprotective activities, indicating that glycosides may be the major hepatoprotective constituents of JH. Besides, only some compounds showed weak antibacterial potentials. These findings supplied a preliminary bioactivity basis for the antioxidant, hepatoprotective, and antibacterial effects of JH. It is also important to understand the chemical basis of the healthcare values of JH flowers and indicated that the compounds can be used for the development of health herb tea and even potential new drugs for the treatment of liver diseases.
Acknowledgments
This work was financially supported by Changjiang Scholars Program in Ministry Education, People’s Republic of China (T2019133), Changsha Municipal Natural Science Foundation (kq2014086), Scientific Research Fund of Hunan University of Chinese Medicine (2021XJJJ006) and Hunan Kangdejia Forestry Technology Co., Ltd., Yongzhou.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Aromatic constituents from the leaves of Actinidia arguta with antioxidant and α-glucosidase inhibitory activity. Antioxidants.. 2021;10:1896-1908.
- [CrossRef] [Google Scholar]
- New roles for cis-jasmone as an insect semiochemical and in plant defense. P. Natl. Acad. Sci. USA. 2000;97:9329-9334.
- [CrossRef] [Google Scholar]
- Flavonoids and caffeoylquinic acids in Chrysanthemum morifolium Ramat flowers: A potentially rich source of bioactive compounds. Food Chem.. 2020;344:128733
- [CrossRef] [Google Scholar]
- The constituents of Eucommia ulmoides OLIV. IV. Isolation of a new sesquilignan glycoside and iridoids. Chem. Pharm. Bull.. 1986;34:4933-4938.
- [CrossRef] [Google Scholar]
- Chemical constituents from Artemisia iwayomogi increase the function of osteoblastic MC3T3-E1 cells. Nat. Prod. Sci.. 2009;15:192-197.
- [Google Scholar]
- Hepatoprotective effect of forsythiaside a against acetaminophen-induced liver injury in zebrafish: Coupling network pharmacology with biochemical pharmacology. J. Ethnopharmacol.. 2021;271:113890
- [CrossRef] [Google Scholar]
- Caffeoylquinic acid derivatives from Inula cappa. Nat. Prod. Res. Dev.. 2014;26:1948-1952.
- [CrossRef] [Google Scholar]
- Bioactive sesquiterpenoids from the rhizomes of Acorus calamus. J. Nat. Prod.. 2012;75:1083-1089.
- [CrossRef] [Google Scholar]
- Chemical constituents of Cercis chinensis leaves. Chem. Nat. Compd.. 2019;55:107-109.
- [CrossRef] [Google Scholar]
- Chemical constituents of Hedyotis diffusa and their anti-inflammatory bioactivities. Antioxidants.. 2022;11:335-349.
- [CrossRef] [Google Scholar]
- One new triterpene ester from Nepeta suavis. J. Asian Nat. Prod. Res.. 2009;11:997-1000.
- [CrossRef] [Google Scholar]
- Flavonoid deactivation of excited state flavins: reaction monitoring by mass spectrometry. J. Agr. Food Chem.. 2012;60:9261-9272.
- [CrossRef] [Google Scholar]
- Two new phenolic glucosides from Lagerstroemia speciosa. Molecules. 2015;20:4483-4491.
- [CrossRef] [Google Scholar]
- Antibacterial bibenzyl derivatives from the tubers of Bletilla striata. Phytochemistry. 2019;162:216-223.
- [CrossRef] [Google Scholar]
- Chemistry and pharmacological activity of sesquiterpenoids from the Chrysanthemum genus. Molecules. 2021;26:3038-3051.
- [CrossRef] [Google Scholar]
- Chemical constituents isolated from the root bark of Cudrania tricuspidata and their potential neuroprotective effects. J. Nat. Prod.. 2016;79:1938-1951.
- [CrossRef] [Google Scholar]
- Identification of phenolic components of chrysanthemum flower (Chrysanthemum morifolium Ramat) Food Chem.. 2010;120:319-326.
- [CrossRef] [Google Scholar]
- Chemical constituents from stem barks of Vernonia cumingiana. China J. Chin. Mater. Med.. 2010;35:1421-1424.
- [CrossRef] [Google Scholar]
- Structural characterization and prebiotic potential of an acidic polysaccharide from Imperial Chrysanthemum. Nat. Prod. Res.. 2020;36:586-1584.
- [CrossRef] [Google Scholar]
- Phenolpropane compounds of Cordia dichotoma and their anticomplementary activities. Chem. Nat. Compd.. 2021;57:169-170.
- [CrossRef] [Google Scholar]
- Solvent effect on phenolics and antioxidant activity of Huangshan Gongju (Dendranthema morifolium (Ramat) Tzvel. cv. Gongju) extract. Food Chem. Toxicol.. 2020;147:111875
- [CrossRef] [Google Scholar]
- Sesquiterpene lactones from Artemisia barrelieri. Phytochemistry. 1991;30:3661-3668.
- [CrossRef] [Google Scholar]
- Phenolic and acetylenic metabolites from Artemisia assoana. Phytochemistry. 1987;26:2619-2624.
- [CrossRef] [Google Scholar]
- (-)-cis-chrysanthenol O-β-D-glucopyranoside: a new monoterpene glucoside from Dicoria canescens. Phytochemistry. 1974;13:2881-2882.
- [CrossRef] [Google Scholar]
- Iridoid glucosides from the fruit of Genipa americana. Chem. Pharma. Bull.. 2005;53:1342-1344.
- [CrossRef] [Google Scholar]
- Biotransformation of (-)-borneol by cultured cells of Eucalyptus perriniana. Phytochemistry. 1993;34:1045-1048.
- [CrossRef] [Google Scholar]
- Monoterpenoid chemistry. Part 3. Stereoselective synthesis of the major oxygenated metabolites of trans-sobrerol. Helv. Chim. Acta.. 1987;70:71-78.
- [CrossRef] [Google Scholar]
- New glycosides from Dracocephalum tanguticum maxim. Arch. Pharm. Res.. 2011;34:2015-2020.
- [CrossRef] [Google Scholar]
- Natural deep eutectic solvent (NADES): A strategy to improve the bioavailability of blueberry phenolic compounds in a ready-to-use extract. Food Chem.. 2021;364:130370
- [CrossRef] [Google Scholar]
- Circular dichroism, a powerful tool for the assessment of absolute configuration of flavonoids. Phytochemistry. 2005;66:2177-2215.
- [CrossRef] [Google Scholar]
- Norisoprenoids from Ulva lactuca. J. Asian Nat. Prod. Res.. 2007;9:321-325.
- [CrossRef] [Google Scholar]
- Transformation of a monoterpene ketone, piperitenone, and related terpenoids using Mucor piriformis. Can. J. Microbiol.. 2005;51:447-454.
- [CrossRef] [Google Scholar]
- Chemical constituents and antifungal activity of Ficus hirta Vahl. fruits. Plants.. 2017;6:44-52.
- [CrossRef] [Google Scholar]
- Cyclohexanecarboxylic-acid derivatives from Psiadia trinervia. Helv. Chim. Acta.. 1992;75:269-275.
- [CrossRef] [Google Scholar]
- Antibacterial and antioxidant flavonoid derivatives from the fruits of Metaplexis japonica. Food Chem.. 2019;289:308-312.
- [CrossRef] [Google Scholar]
- Chemical constituents from Buddleja officinalis and their inhibitory effects on nitric oxide production. Nat. Prod. Sci.. 2016;22:129-133.
- [CrossRef] [Google Scholar]
- Neuroprotective caffeoylquinic acid derivatives from the flowers of Chrysanthemum morifolium. J. Nat. Prod.. 2017;80:1028-1033.
- [CrossRef] [Google Scholar]
- Phenolics from strawberry cv. Falandi and their antioxidant and α-glucosidase inhibitory activities. Food Chem.. 2016;194:857-863.
- [CrossRef] [Google Scholar]
- Six new compounds from the flowers of Chrysanthemum morifolium and their biological activities. Bioorg. Chem.. 2019;82:139-144.
- [CrossRef] [Google Scholar]
- The flower head of Chrysanthemum morifolium Ramat. (Juhua): A paradigm of flowers serving as Chinese dietary herbal medicine. J. Ethnopharmacol.. 2020;261:113043
- [CrossRef] [Google Scholar]
- Structural analysis and antioxidant activity of the glycoside from Imperial Chrysanthemum. Bioorg. Med. Chem. Lett.. 2018;28:1581-1590.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.104292.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







