Translate this page into:
Structure based approach for twin-enzyme targeted benzimidazolyl-1,2,4-triazole molecular hybrids as antifungal agents
⁎Corresponding authors. radha-cm@pau.edu (Radha Ahuja), anjalisidhu@pau.edu (Anjali Sidhu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Agri-vitality of benzimidazoles and 1,2,4-triazoles against ergosterol and β-Tubulin prompted and “lead hybridization” based novel series of benzimidazolyl-1,2,4-triazoles (1–32) were designed as twin-enzyme targeted inhibitors. In silico computational tools viz. molecular docking, Lipinski parameters, Frontier molecular orbital approach and Toxicity analysis screened three benzimidazoly-1,2,4-triazoles out of 32 designed molecules, which were synthesised by multistep protocol and characterized by spectroscopic techniques. Antimycotic activity against F. verticillioides, D. oryzae, C. lunata and F. fujikuroi indicated deca fold enhanced potency of all the synthesised compounds, than the standard commercial benzimidazole fungicide, carbendazim. Compounds 8 exhibited ED50 values lower than triazole fungicide, propiconazole. Remarkably, compound 8 inflicted the most promising activity against all the test fungi with ED50 value ranging from 16 to 21 μg/ml better than the standard commercial fungicides used (Tilt: 20–25 μg/ml and Carbendazim:150–230 μg/ml). Ultra microscopic details revealed compound 8 not only caused aberrant distortions resulting in collapsed hyphae but also efficiently shrunken the spores resulting in reproduction inhibition, as possible cause of fungal growth inhibition.
Keywords
Benzimidazole
Docking
Lanosterol 14α-demethylase
Tubulin
1,2,4-Triazole
1 Introduction
Benzimidazoles, potent β-tubulin polymerization inhibitors in many species of fungi are used as phyto-fungal control agents since the late 1960s. Its commercial analogous viz. carbendazim, benomyl, thiabendazole, mebendazole and fubridazole are effective against fungal pathogens of cereals, fruits, vegetables and vines. Fungicidal applications are facing threat because of their single site mode of action (https://en.wikipedia.org/wiki/Benzimidazole_fungicide) making their phytoapplication more prone to collateral crop damage upon development of fungicidal resistance. Limitations of toxicity (Mandapati et al., 2014) and development of resistance demand the exploration of novel benzimidazole analogous with strong activity, low toxicity and with multisite mode of action.
1,2,4-triazoles are of great interest to agriculturalists owing to their ubiquitous worth in pathological system. Its analogues viz. cyproconazole, propiconazole, azaconazole, febuconazole and tebuconazole, commercial fungicides are considered as bioisosteres of imadazole (Rezaei et al., 2009). It is a momentous structural moiety for antifungal activity. N-4 of triazole binds to heme iron, thereby, inhibiting the action of 14α-demethylase in biosynthesis of ergosterol in fungus. It can capably interact with various active targets in biological system via diverse non-covalent interactions like hydrogen bonds, coordination, ion dipole, п-п stacking and hydrophobic effect as well as Van der Waals force (Wang et al., 2012).
Molecular hybridization of bioactive leads in a single molecule is hypothesized as a fruitful strategy providing new chemical entities for hyperactive and multitargeted action (Sidhu and Kukreja, 2015). Hybrids of benzimidazole with 1,2,4-triazole have glimpses in literature regarding their positive outcomes on biopotential. Can et al., (2017) reported anticandidal properties of triazole-based benzimidazoles as lanesterol 14α-demethylase inhibitors with MIC50 values between 0.78 and 1.56 μg/ml. Benzimidazole based fluconazole exhibited enhanced binding ability with active sites of lanesterol 14α-demethylase and hence antifungal potential against Candida albicans with MIC of 0.0075 μmol/ml (Kankate et al., 2019). Chidambaranathan and Mahalakshmi (2015) have reported another permutation of these two bioactive moieties inhibiting E. coli at MIC of 4 μg/ml. Apart from antifungal action, this combination has also been employed for the treatment of non-small cell lung cancer (Bistrovic et al., 2018). Benzimidazol-2-yl-1,2,4-triazol have not yet been explored to manage the fungal infestations on agriscenario.
In silico screening of molecules have great potential to speed the rate of discovery while reducing the need for expensive lab work and in vivo trials. Virtual screening has become an integral part of the lead discovery process. Computer Aided Drug Designing (CADD) methods can surge the probabilities of recognizing compounds with desirable characteristics, hustle up the hit-to-lead development, and expand the odds of getting a compound over the many obstacles of pre-testing. Overall, in silico efforts are a push towards green chemistry approach.
The present paper presents the designing of novel benzimidazolyl-1,2,4-triazoles having variety of spacers between the two leads, followed by in silico filtration to get the high potential mycocidal molecules with multisite mode of action. The screened molecules were synthesized and evaluated for their antifungal potential against various phytopathogenic fungi of rice. The ultramicroscopic details described morphological alteration as possible cause of fungal inhibition.
2 Materials and methods
All the solvents and reagents were commercially available high grade materials. The products were splendidly purified and the purity of all the compounds was checked on a silica gel-G plates. Melting points of the synthesized derivatives were determined in open capillary tubes and were uncorrected. The IR spectra were recorded on Bruker Alpha-2 in our laboratory, PAU, Ludhiana. 1H NMR and mass spectroscopic analysis were recorded in Sophisticated Analytical Instrument Facility, Panjab University Chandigarh, India. 1H NMR spectra were recorded on a Bruker Avance II 400 NMR spectrometer with DMSO or CDCl3 as solvents and TMS as internal standards. The chemical shifts were expressed in δ (ppm) values. Mass spectra were recorded by using Waters Q-TOF LC-MS.
2.1 Rationale of designing of benzimidazolyl-1,2,4-triazoles
“Aka targeted” fungicides like benzimidazoles and 1,2,4-triazoles act at specific point in a biosynthetic pathway, rendering them less effective or ineffective on any alteration on the same point in the pathogen. Such molecules with single site mode of action are more prone to the development of fungicidal resistance, which is an evolutionary process and a stable heritable trait. The use of alternate fungicides with same FRAC codes does not work as they share similar mode of action (Vincelli 2014). FRAC codes that include “M” (Multi-site Action) is the desirable trait and are generally the hotspot of discovery of novel molecules of present day pesticidal research.
Maintaining the functional integrity of individual bioactive leads, “lead hybridization” strategy is continuously explored to get new molecular entities with “M” status of their mode of action. 1,2,4-Triazole, a well known antifungal scaffold inhibits ergosterol biosynthesis by binding to Cyp51 family of cytochrome P450 (George et al., 2014). Benzimidazoles, another important category of agri-fungicides, act by binding to β-tubulin preventing their polymerization into fungal microtubules, thus, preventing hyphal growth (https://en.wikipedia.org/wiki/Benzimidazole). Molecular hybrids of two bioactive moieties act on different points of fungal metabolic pathways, thus, inhibiting the fungal growth in synergistic manner, with less chances of resistance development.
The work is designed with rationale of in silico screening and optimization of the novel benzimidazole decorated 1,2,4-triazole separated by spacers with best twin-docking into β-tubulin and lanosterol 14α-demethylase enzymes along with most favoured physicochemical, FMO parameters and toxicity factors for their best anticipated antifungal activity
2.2 In silico studies
2.2.1 Docking studies
Twin-enzyme docking strategy was employed to evaluate the binding mode of compounds into active site of tubulin and lanosterol 14α-demethylase separately, using AutoDockTools-1.5.6 software. Crystallographic full-length lanosterol 14 alpha-demethylases of prominent fungal pathogens candida albicans protein (PDB: 5V5Z) and yeast tubulin (PDB: 5W3F), were retrieved from the Protein Data Bank (PDB) (http://www.rcsb.org). The enzymes were made to bind with compound and docking score was calculated. The more negative energy represented the effective binding and hence activity of compound.
2.2.1.1 Ligand preparation
The structures of compounds were saved in mol format using ChemDraw Ultra 8.0. Propiconazole and Carbendazim were chosen as references. The compounds were optimized by program Chem3D Ultra 8.0 using molecular dynamics MM2 algorithm and saved as PDB-files. Molecular mechanics were used to produce more realistic geometry values for the majority of synthesized organic molecules owing to the fact of being highly parameterized. The pdb-files were converted to PDBQT By using AutoDockTools-1.5.6 (Trott and Olson, 2009).
2.2.1.2 Protein preparation
The water molecules and ligand from the crystals (in PDB files) were deleted by Discovery Studio 4.0. The proteins were saved as pdb-files. In AutoDockTools-1.5.7, polar hydrogens were added and saved as PDBQT. Grid generation was done by removing of previously bound inhibitor at active site of X-ray crystal structure. The grid size was generated for serving active site protein structures of 5V5Z and 5W3F. Grid box for 5V5Z was set as follows: center_x = 24.263, center_y = 43.676, center_z = 31.008, size_x = 70, size_y = 70, size_z = 70 and for 5W3F at center_x = 308.282, center_y = 436.754, center_z = 296, size_x = 126, size_y = 86, size_z = 86. Vina was used to carry out docking. For visualization Biovia Discovery Studio Visualizer (v17.2.0.16349, Accelrys, San Diego, CA, USA) was applied.
2.2.1.3 Ligand-protein interactions
The automatic ligand-protein interactions diagrams were generated from standard PDB file input using LigPlot software (Laskowski and Swindells, 2011). The interactions generated provide information about hydrogen bonds and the hydrophobic contacts. The indication of hydrogen bonds is done by dashed lines between the atoms involved, while hydrophobic contacts are indicated by an arc with spokes radiating towards the atoms of ligands they contact.
2.2.2 Lipinski filtration
Lipinski's parameters of designed series were calculated using Molinspiration, web-based software, which determine the degree of permeability of compounds through biomembranes. Good bioavailability of compounds will satisfy the Lipinski rule where the maximum molecular weight of the compound is 500, logP is not greater than 5, the number of hydrogen bond donor is less than 5, and hydrogen bond acceptor is less than 10 (Lipinski et al., 2012). logP is logarithm of the partition coefficient in n-octanol/water system that reflects the overall lipophilicity of a molecule, a property of major importance in biochemical applications, the molecular weight defines the effect of size of the molecule on its biological activity and the number of hydrogen donors (nOHNH) and hydrogen acceptors (nON) determine the upper limits for the biomolecules to be able to penetrate through biomembranes. Molecules that would obey these rules should exert acceptable solubility and cell permeability properties and were defined as ‘pesticide-like’ (Tice, 2001).
2.2.3 Frontier molecular orbital energy
Molecular mechanics approach using Chem3D Ultra 8.0 was used for the calculations of frontier molecular orbital energies to predict the toxicity of the compounds. The energy gap between HOMO (highest occupied molecular orbital) and LUMO (lowest unoccupied molecular orbital) give information regarding binding modes of the molecules.
2.2.4 Toxtree analysis
Toxtree v.2.6.6 estimates toxic hazards of chemicals and divides them into classes of toxicity profile. The software predicts toxic effect by applying various decision-tree approaches. In order to measure toxic hazards of all the synthesized compounds two dimensional models of all compounds were converted into SMILE format (www.molinspiration.com) using online SMILE convertor which were then put into chemical identifier row available in software to get toxicity parameters (Weininger, 1988).
2.3 Experimental procedures
2.3.1 General synthetic procedure for the preparation of 4-(1H-benzo[d]imidazol-2-yl)aryl 2-(1H-1,2,4-triazol-1-yl)acetate (1 and 4)
1a-1b (10 mmol) and piperazine (0.5 g) dissolved in acetonitrile (20 ml) were stirred at room temperature for 5 min. Chloroacetyl chloride (15 mmol) was added drop wise to this solution under vigorous stirring at room temperature. After completion of reaction 1H-1,2,4-triazole (10 mmol) was added to the reaction mixture and stirring was continued till completion of reaction. The intermediates 1c-1d were obtained by evaporating the solvent under reduced pressure. Equimolar amounts of o-phenylendiamine (10 mmol) and intermediates 1c-1d (10 mmol) were dissolved in water (50 ml), sodium bisulfite (0.4 g) was added and the mixture was stirred vigorously at 80 °C till the completion of reaction. The mixture was cooled to room temperature and product separated was filtered, washed with water several times and air dried.
2.3.1.1 4-(1H-benzo[d]imidazol-2-yl)phenyl 2-(1H-1,2,4-triazol-1-yl)acetate (1)
Yield: 60%, mp: 243–245 °C, FTIR υ (ATR, cm−1): 3377 (NH), 1731 (C⚌O), 1158 (C—N), 1382, 1515 (C⚌N), δ: 9.39 (s, 1H, NH, Benzimidazole), 8.45 (s, 2H, triazole), 6.56–7.65 (m, 8H, Ar—H), 4.23 (s, 2H, CH2), 13C NMR (DMSO‑d6): δ (ppm): 20.82, 31.52, 41.27, 113.56, 124.54, 126.18, 128.12, 128.75, 129.22, 129.37, 130.91, 134.26, 135.73, 150.37, 151.08, 155.30, 190.85. MS (ESI): m/z = 317.14
2.3.1.2 4-(1H-benzo[d]imidazol-2-yl)-2-methoxyphenyl 2-(1H-1,2,4-triazol-1-yl)acetate (4)
Yield: 53%, mp: 208–210 °C, FTIR υ (ATR, cm−1): 3367 (NH), 1734 (C⚌O), 1159 (C—N), 1383 and 1515 (C⚌N). 1H NMR (400 MHz, DMSO‑d6) δ: 9.72 (s, 1H, NH), 8.26 (s, 2H, triazole), 6.62–7.81 (m, 8H, Ar—H), 3.17 (s, 2H, CH2), 13C NMR (DMSO‑d6): δ (ppm): 21.71, 32.49, 41.29, 53.22, 115.76, 124.54, 127.18, 128.42, 128.93, 129.31, 131.62, 132.37, 134.26, 135.73, 150.37, 151.08, 155.30, 191.85. MS (ESI): m/z = 349.11
2.3.2 Synthetic procedure for the preparation of 4-(1H-benzo[d]imidazol-2-yl)-2-methoxyphenyl 2-(1H-1,2,4-triazol-1-ylamino)acetate (8)
To a stirring solution of 1b (10 mmol) and piperazine (0.5 g) in acetonitrile (20 ml), chloroacetyl chloride (15 mmol) was added drop wise at room temperature. After completion of reaction, 4-amino-1, 2, 4-triazole (10 mmol) was added to the reaction mixture. The precipitates of intermediates 1e separated were collected, washed with acetonitrile and air dried. Equimolar amounts of o-phenylenediamine (10 mmol) and intermediate 1e (10 mmol) was dissolved in water (50 ml), sodium bisulfite (0.4 g) was added and the mixture was stirred vigorously at 80 °C till the completion of reaction. The mixture was cooled to room temperature and product separated was filtered, washed with water several times and air dried.
Yield: 58%, mp: 224–226 °C, FTIR υ (ATR, cm−1): 3307, 3261 (NH), 1738 (C⚌O), 1027, 1157 (C—N), 1462, 1510 (C⚌N), 1H NMR (500 MHz, CDCl3) δ: 8.92 (s, 1H, NH, Benzimidazole), 7.86 (s, 2H, triazole), 6.85–7.61 (m, 7H, Ar—H), 3.81 (s, 3H, OCH3), 3.70–3.73 (d, 2H, CH2), 1.73–1.76 (t, 1H, NH), 13C NMR (DMSO‑d6): δ (ppm): 25.23, 33.25, 49.27, 55.32 , 115.55, 126.83, 127.68, 128.47, 128.85, 129.22, 129.58, 130.37, 131.33, 132.35, 134.76, 152.54, 154.30, 192.85. MS (ESI): m/z = 364.17
2.4 Antifungal evaluation studies of synthesized compounds
The synthesized compounds were nano-formulated using sonochemical method (Sidhu and Kukreja, 2015) by dissolving them (25 mg) in 1 ml Triton-X under sonication and making total volume of 50 ml with sterilized water. The solution was stored as a stock of 500 μg/ml.
2.4.1 Test fungi
Four pathogenic fungi of rice viz. F. verticillioides, D. oryzae, C. lunata and F. fujikuroi were isolated from discoloured rice seeds using the standard blotter method of detection of seed borne pathogens (ISTA, 1993). The fungi were identified by morphological (Booth, 1977) and PCR technique. F. verticillioides was identified using VER1 (CTTCCTGCGATGTTTCTCC) and VER2 (AATTGGCCATTGGTATTATATATCTA) primers for the amplification of translation elongation factor 1-α gene region of DNA (Faria et al., 2011; Mule et al., 2004). The amplification of ITS genes was done using primers PN3 (CGTTGGTGAACCAGCGGAGGGATC) and PN16 (TCCCTTTCAACAATTTCACG) (Neuvéglise et al., 1994; Sunpapao et al., 2014) for identification of C. lunata. D. oryzae was identified by amplification of ITS regions using primers ITS1 (TCCGTAGCTGAACCTGCCG) and ITS4 (TCCTCCGCTTATTGATATGC) (Weikert-Oliveira et al., 2002). For identification of F. fujikuroi, EF-1α gene was amplified using primers EF1 (5′-ATGGGTAAGGA (A⁄G)GACAAGA C-3′) and EF2 (5′- GGA(G/A)GTACCAGT(G/C)AT CATGTT-3′) (Hsuan et al., 2011). The pure cultures were maintained on potato dextrose agar (PDA) medium slants at 4 °C.
The synthesized compounds (1, 4 and 8) were tested at different concentrations of 10, 20, 50 and 100 μg/ml against test fungi using poisoned food technique (Nene and Thapliyal, 1997). For comparative evaluation, propiconazole (1-[[2-(2,4-dichlorophenyl)-4-propyl-1,3-dioxolan-2-yl]methyl]-1H-1,2,4-triazole) (10, 20, 50 and 100 μg/ml) and carbendazim (Methyl 1H-benzimidazol-2-ylcarbamate) (100, 200, 300, 400 and 500 μg/ml) were used as standard fungicides. Antifungal results were expressed in terms of ED50 values.
2.5 SEM analysis
Fungal culture was fixed in 2.5% cacodylate buffered glutaraldehyde at 4 °C for 24 h, washed thrice with 0.1 M cacodylate buffer at 4 °C and fixed with 1% osmium tetraoxide at 4 °C for 2 h. A dehydration series was carried out with 30, 50 and 70% ethanol for 15 min at 4 °C and 80% and 90% at room temperature. The dried specimens were placed on conductive carbon tabs on an aluminium stub. The samples were imaged by Hitachi S-3400 N SEM at 15 kV.
2.6 Statistical analysis
The triplicate percent inhibition data of antifungal evaluation studies were analyzed by Probit analysis to determine ED50 values using IBM SPSS 23 software (Finney, 1971).
3 Results and discussion
3.1 In silico guided virtual screening of designed hybrids
A series of 32 compounds containing benzimidazole and 1,2,4-triazole, in a single molecule, were designed using variety of spacers between the two bioactive moieties. Various computational tools were employed to check their in silico efficacy for their potential use as antifungal compounds. AutoDockTools evaluated their binding into the active sites of β-tubulin and lanosterol 14α-demethylase (CYP51). The best compounds were investigated for their mode of interactions into the active sites via LigPlot+. Deviations from favourable physicochemical molecular descriptors were done using Lipinski filtration. Toxtree analysis along with FMO approach gave us the idea about potential toxicities. Various steps of virtual screening of the designed molecules are given in Fig. 1.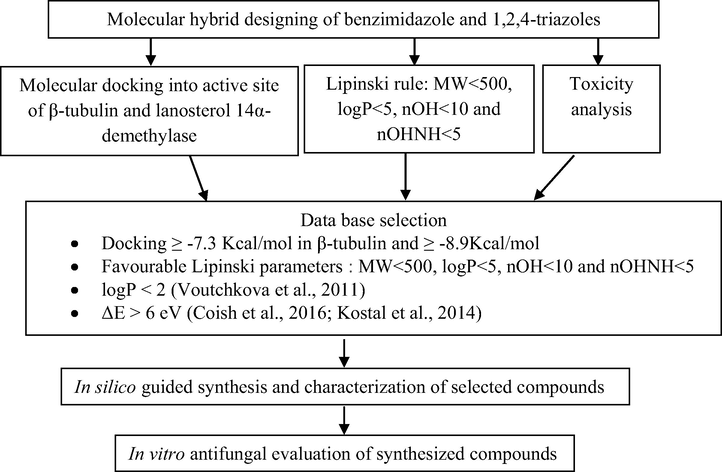
Virtual screening campaign used in selection of the most potent compounds.
3.1.1 Molecular docking studies
Benzimidazoles are myco-inhibitors via tubulin protein polymerization inhibition by binding to β-tubulin while 1,2,4-triazoles are well known lanosterol 14α-demethylas inhibitors (Aoyama et al., 1984; Kelly et al., 1993). The designed molecules were investigated for their best co-inhibiting activities into these two enzymes with reference to commercial fungicides (carbendazim and propiconazole). Therefore, molecular docking of designed hybrids were carried out into the active site of the tubulin and lanosterol 14α-demethylase using AutoDockTools-1.5.6. Lanosterol 14 alpha-demethylase of prominent fungal pathogens candida albicans protein (PDB: 5V5Z) and yeast tubulin (PDB: 5W3F) were used.
All the designed 32 compounds fitted well into the binding pocket of β-tubulin and showed appreciable docking score (between −6.1 and −9.3 Kcal/mol) in comparison to the standard carbendazim (Methyl 1H-benzimidazol-2-ylcarbamate) (−5.6 Kcal/mol). The median of docking scores of the designed series was approximately −7.3 Kcal/mol. Out of 32 compounds, twelve compounds (1–2, 4, 6, 8–12, 22, 25 and 30) showed docking scores ≥ -7.3 Kcal/mol. The docking score in the active site of lanosterol 14α-demethylase of the compounds were between −7.8 Kcal/mol and −9.4 Kcal/mol (Table 1) which were better than that of standard propiconazole (1-[[2-(2,4-dichlorophenyl)-4-propyl-1,3-dioxolan-2-yl]methyl]-1H-1,2,4-triazole) (-7.2Kcal/mol). Eleven compounds viz. 1, 4–5, 8–11, 13, 25, 28 and 30 showed appreciable docking score ≥−8.9Kcal/mol i.e median of all the values, in this binding pocket. Eight compounds viz. 1, 4, 8–11, 25 and 30 were selected based on their most significant docking scores (more than median of all the docking scores) into lanosterol 14α-demethylase which also fitted well into β-tubulin. logP = logarithm of octanal/water partition coefficient; MW = molecular weight; nON = number of H-bond acceptor; nOHNH = number of H-bond donors.
Compound
R
R′
Docking score (Kcal/mol)
logP
MW
n ON
n OHNH
Tubulin
Lanosterol 14α-demethylase
1
—H
6-COCH2—
−7.3
−9.0
1.94
319
7
1
2
—H
5-COCH2—
−7.4
−8.7
2.01
319
7
1
3
—H
4-COCH2—
−6.8
−8.2
1.99
319
7
1
4
—OCH3
6-COCH2—
−7.4
−8.9
1.70
349
8
1
5
—H
6-COCH2NH—
−7.2
−8.9
1.79
334
8
2
6
—H
5-COCH2NH—
−7.3
−8.7
1.77
334
8
2
7
—H
4-COCH2NH—
−6.9
−7.8
1.75
334
8
2
8
—OCH3
6-COCH2NH—
−9.3
−8.9
1.38
364
9
2
9
—H
6-CH2CO—
−7.4
−9.4
2.63
319
7
1
10
—H
5- CH2CO—
−7.9
−9.3
2.60
319
7
1
11
—H
4- CH2CO—
−7.5
−8.9
2.58
319
7
1
12
—OCH3
6- CH2CO—
−8.0
−8.8
1.62
349
8
1
13
—H
6-CH2CONH—
−7.2
−9.1
1.83
334
8
2
14
—H
5-CH2CONH—
−7.1
−8.8
1.81
334
8
2
15
—H
4-CH2CONH—
−6.9
−8.7
1.78
334
8
2
16
—OCH3
6-CH2CONH—
−6.6
−8.6
1.38
364
9
2
17
—H
6-CH2CH2CH2—
−6.7
−8.8
3.02
319
6
1
18
—H
5-CH2CH2CH2—
−6.9
−8.6
3.00
319
6
1
19
—H
4-CH2CH2CH2—
−6.6
−8.1
2.99
319
6
1
20
—OCH3
6-CH2CH2CH2—
−6.1
−7.9
2.61
349
7
1
21
—H
6-CH2CH2CH2NH—
−7.0
−8.5
2.78
334
7
2
22
—H
5-CH2CH2CH2NH—
−7.3
−8.4
2.75
334
7
2
23
—H
4-CH2CH2CH2NH—
−6.8
−8.1
2.70
334
7
2
24
—OCH3
6-CH2CH2CH2NH—
−6.7
−8.3
2.37
364
8
2
25
—H
6-SO—
−8.5
−8.9
2.66
325
7
1
26
—H
5-SO—
−6.4
−8.3
2.62
325
7
1
27
—H
4-SO—
−6.8
−8.4
2.59
325
7
1
28
—OCH3
6-SO—
−6.2
−8.9
2.45
355
8
1
29
—H
6-SONH—
−7.1
−8.8
2.42
340
8
2
30
—H
5-SONH—
−7.6
−8.9
2.40
340
8
2
31
—H
4-SONH—
−6.9
−8.1
2.39
340
8
2
32
—OCH3
6-SONH—
−6.0.8
−8.8
2.30
370
9
2
Carbendazim
–
–
−5.6
–
1.29
191
5
0
Propaconazole
–
–
–
7.2
4.16
342
5
0
Overall, among the 32 compounds designed with various spacers, all results were in favour of presence of —COCH2, COCH2NH—, CH2CO and —CH2CONH— i.e carbonyl group between benzimidazole and 1,2,4-triazole/4-amino-1,2,4-triazole. For first twelve compounds, presence of –OCH3 group was favoured in the docking into active pocket of β-tubulin. In case of docking in lanosterol 14α-demethylase, space at p-position was favoured in comparison to o- and m-position.
3.1.2 Bioavailability
Chemo-informatic properties (Lipinski descriptors) exposed that all the design compounds 1–32 qualified the limits of descriptors logP (<5), molecular weight (<500 g/mol), hydrogen bond acceptor (<10) and donor (<5) (Table 1) i.e all the compounds obeyed all the molecular descriptors under Lipinski’s rule.
logP provides vital information about toxicity of synthetic compounds. The compounds having logP value <2 are likely to have low acute toxicity (Voutchkova et al., 2011; Coish et al., 2016; Kostal et al., 2014). log P < 2 was taken as criteria for screening of designed molecules with hypotoxic effects. Docking pose screening followed by logP < 2 screening qualified only three compounds viz. 1, 4 and 8 out of previously selected best docked eight compounds viz. 1, 4, 8–11, 25 and 30, with best predicted bioactivity and low toxicity.
3.1.3 Toxticity analysis
Toxicity analysis was carried out by FMO approach by calculating HOMO-LUMO energy gap (ΔE) which determines the stability and toxicity patterns of compounds. The safer chemical space to minimize aquatic toxicity has been defined by ΔE > 6 eV (Coish et al., 2016; Kostal et al., 2014). Relatively higher values (7.630–7.656 eV) were recorded for the compounds 1, 4 and 8 (Table 2). Toxicity estimation of FMO approach were in agreement with those of Toxtree analysis indicating that all the compounds belong to class III of toxicity and had persistent biodegradability with no carcinogenicity, which was same as that of carbendazim and propiconazole.
Compound
HOMO (eV)
LUMO (eV)
Energy gap (eV)
1
−8.688
−1.0421
7.645
4
−8.629
−0.999
7.630
8
−8.635
−0.970
7.665
Carbendazim
−8.542
−0.266
8.276
Propiconazole
−5.588
−0.681
4.907
3.1.4 Binding modes of selected three compounds
Rationale of best binding of compounds 1, 4 and 8 into active site of both enzymes were revealed in terms of their H-bonds formation into active pocket of both the enzymes. Only three compounds viz. 1, 4 and 8 were chosen to study their binding modes with targeted enzymes. Binding modes of the selected compounds with targeted enzymes were studied using LigPlot+.
Hydrogen bonding interactions were more pronounced in the selected compounds (as indicated from ligand interaction diagrams) in the binding pockets of both the target enzymes, in comparison to standards (Figs. 2–9). In β-tubulin, the ligand interaction diagram showed more no. of H-bonds (two - five) in all the selected compounds than carbendazim (three) (Table 3). Compounds 1 and 4 exhibited ligand-enzyme interactions via four and five H-bonds with various amino acids mentioned in Table 3, while carbendazim exhibited three H-bonds (Fig. 9). In compound 8, two H-bonds were formed with Gln11 present in binding cleft of active pocket of β-tubulin. In all the three compounds, the right position of this N-atoms of both 1,2,4-triazole and benzimidazole, that O-atoms of methoxy and carbonyl groups, contributed well to strong binding into active site of enzymes via H-bonding.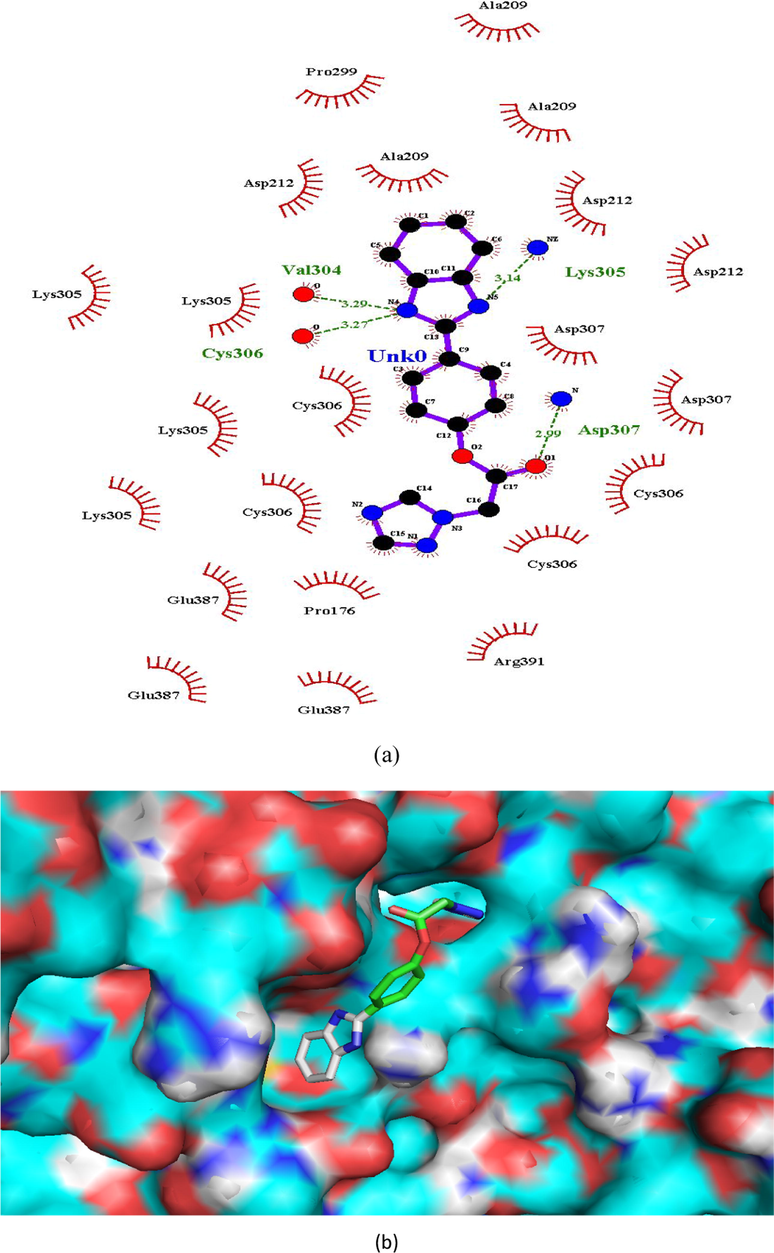
(a and b). Ligand interaction diagram of compound 1 in β-tubulin (a) 2D and (b) 3D view.
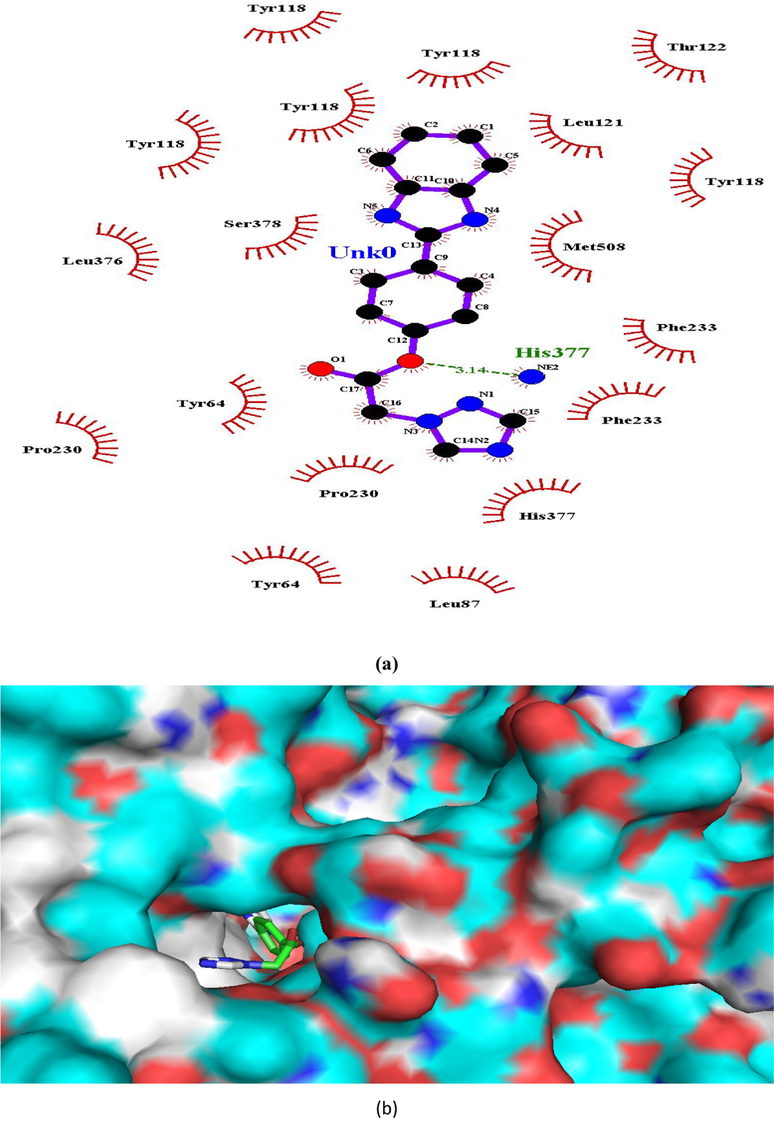
(a and b). Ligand interaction diagram of compound 1 in lanosterol 14α-demethylase (a) 2D and (b) 3D view.
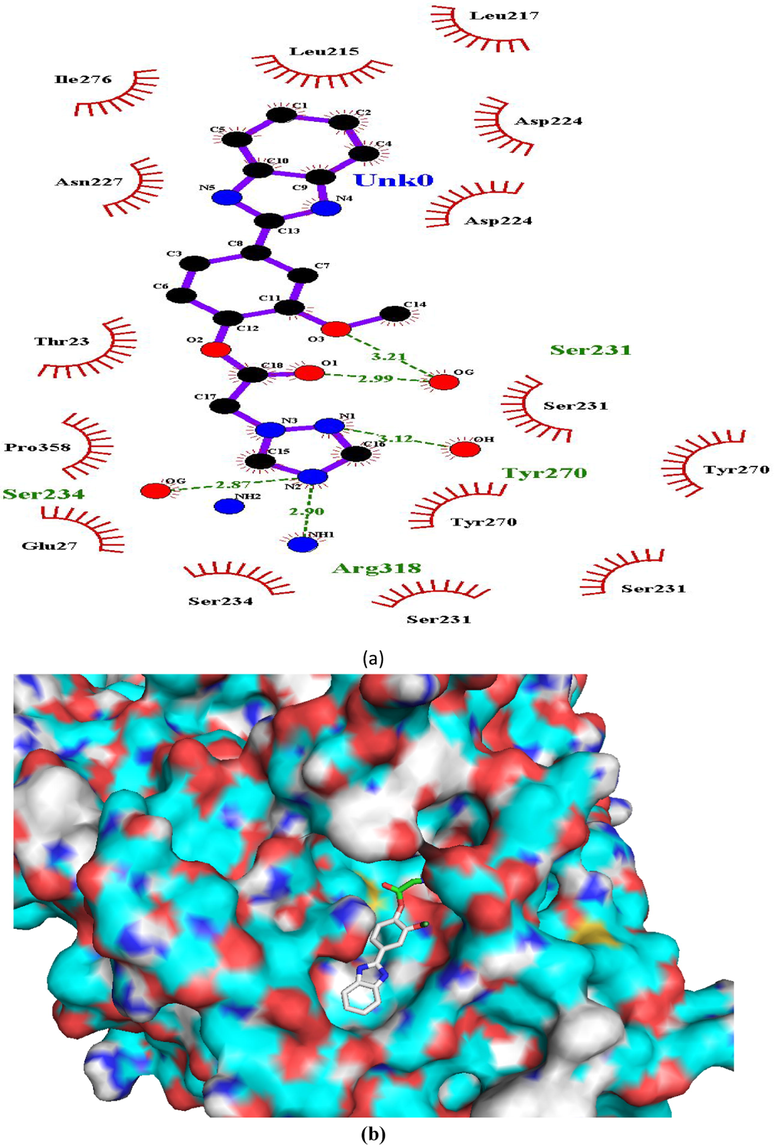
Ligand interaction diagram of compound 4 in β-tubulin (a) 2D and (b) 3D view.
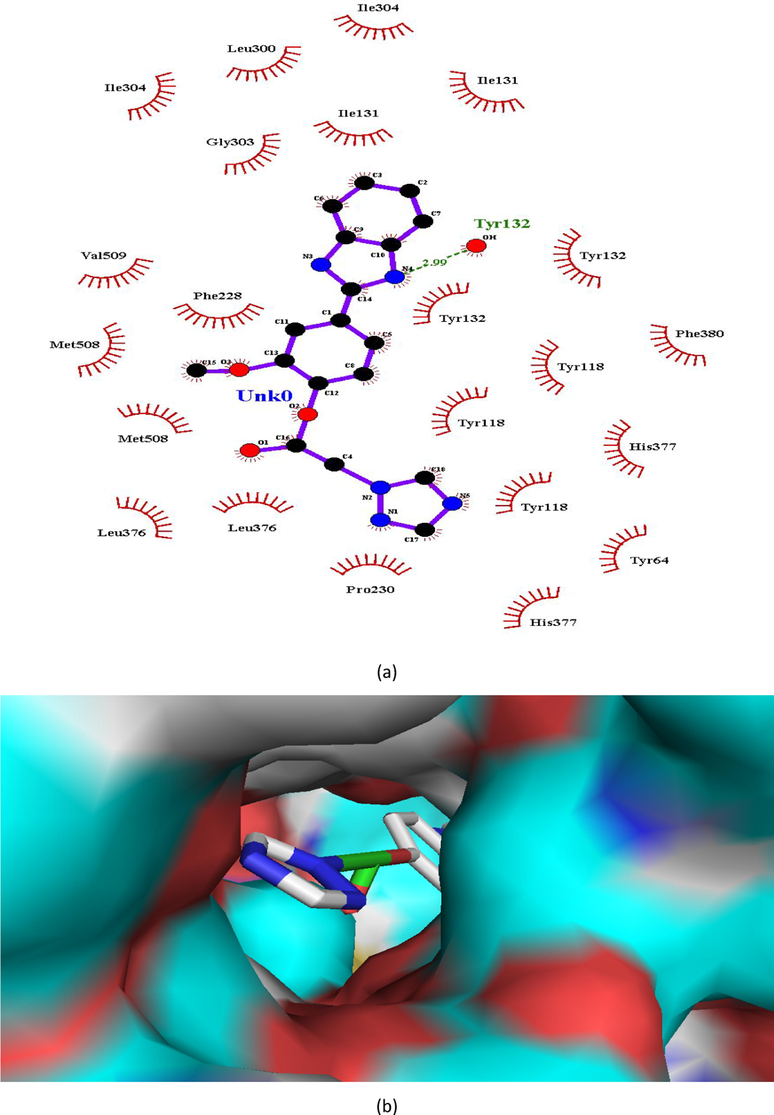
(a and b). Ligand interaction diagram of compound 4 in lanosterol 14α-demethylase (a) 2D and (b) 3D view.
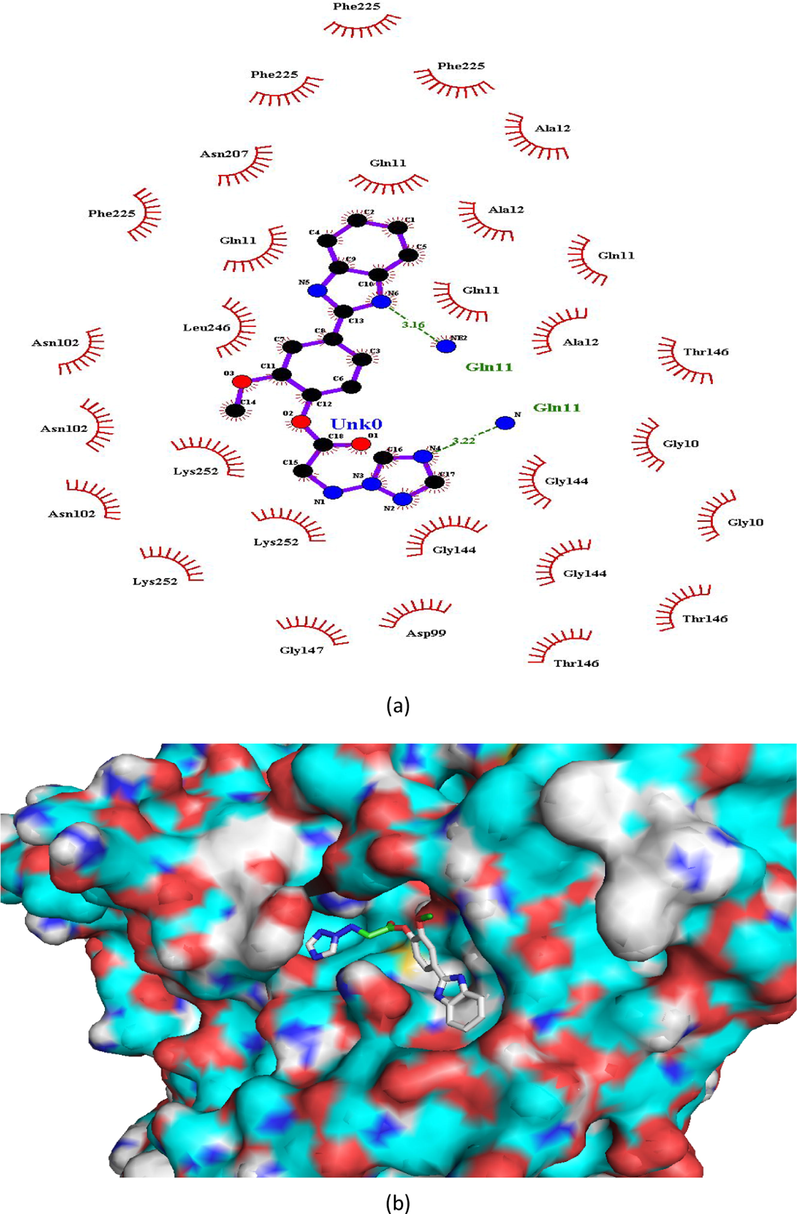
(a and b). Ligand interaction diagram of compound 8 in β-tubulin (a) 2D and (b) 3D view.
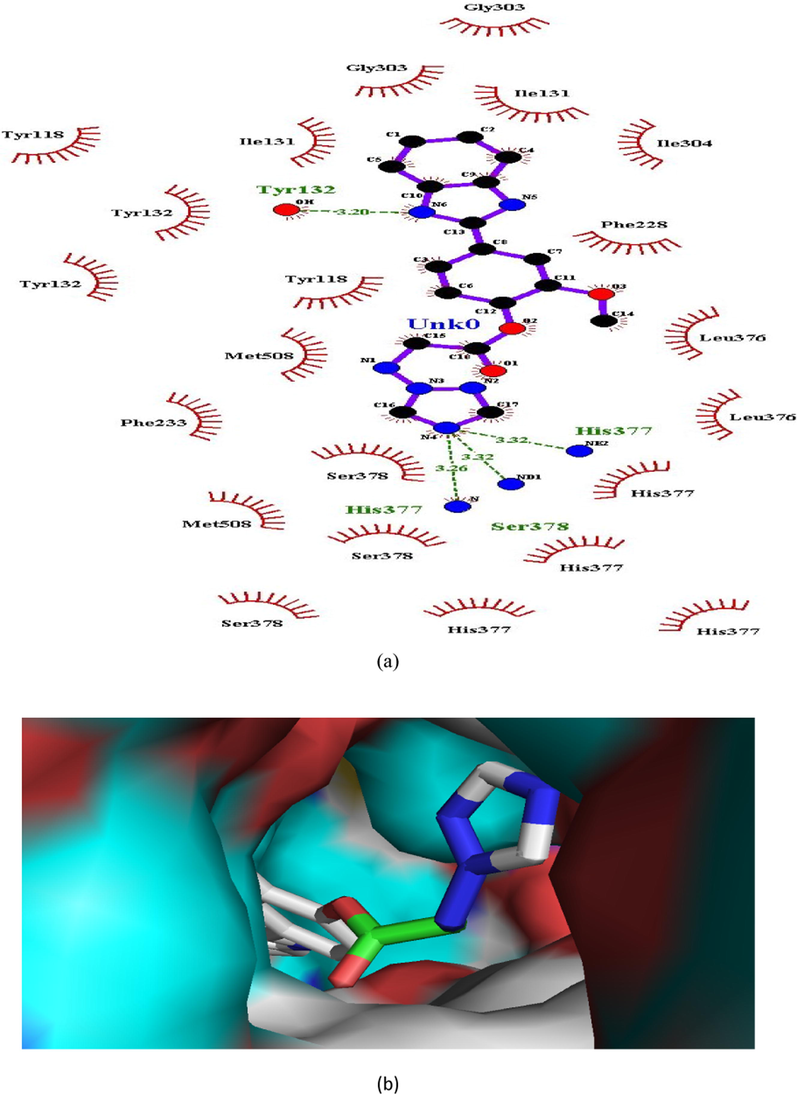
(a and b). Ligand interaction diagram of compound 8 in in lanosterol 14α-demethylase (a) 2D and (b) 3D view.
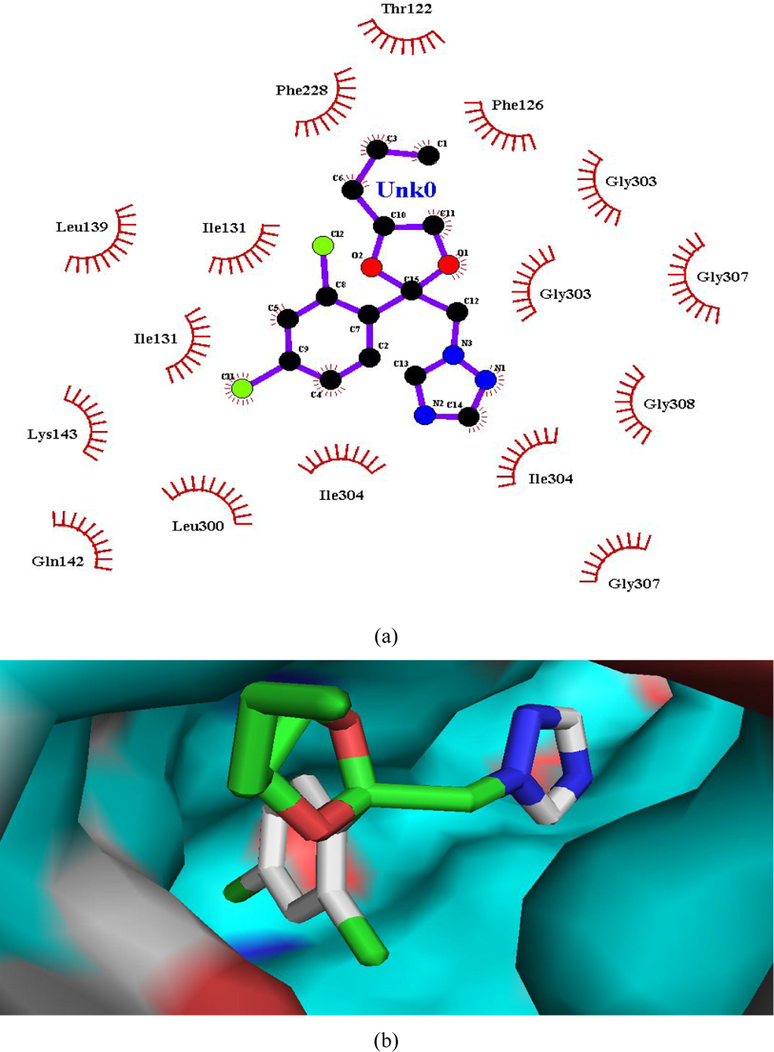
(a and b). Ligand interaction diagram of propiconazole in lanosterol 14α-demethylase (a) 2D and (b) 3D view.
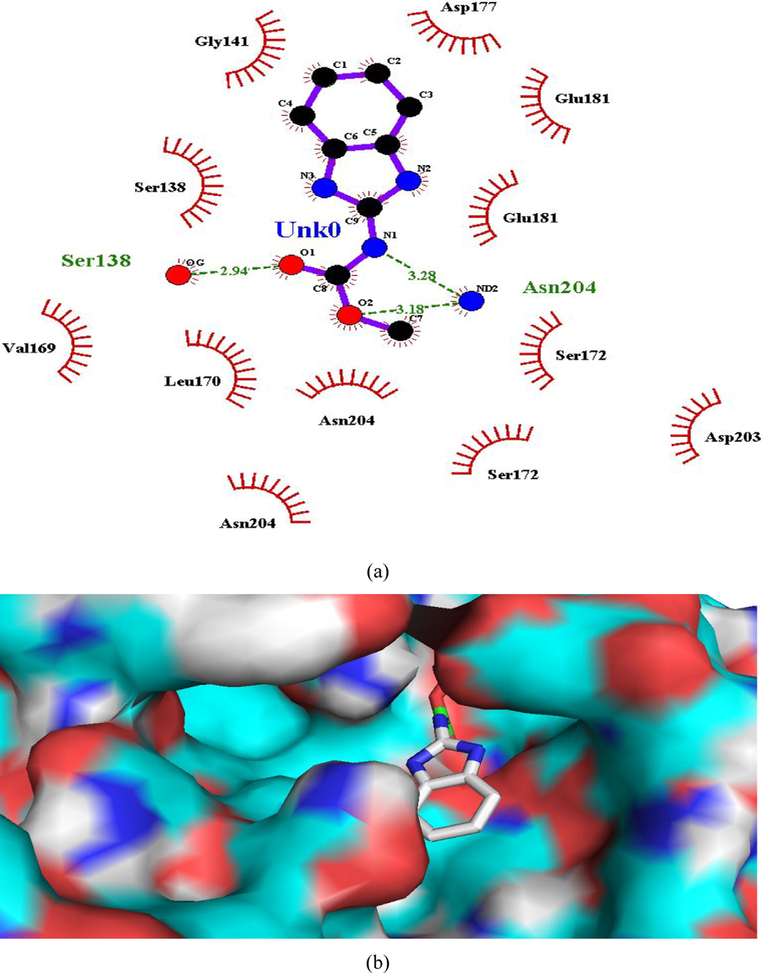
(a and b). Ligand interaction diagram of carbendazim in β-tubulin (a) 2D and (b) 3D view.
Compound
β-Tubulin
Lanosterol 14α-demethylase
H-bonds
Amino acid residues involved
H-bonds
Amino acid residues involved
1
4
Val304, Lys305, Cys306 and Asp307
1
His377
4
5
Ser321, Tyr270, Ars318 and Ser234
1
Tyr132
8
2
Gln11
4
His377, Ser377 and Tyr132
Carbendazim
3
Asn204 and Ser138
–
–
Propiconazole
–
–
0
–
In case of lanosterol 14α-demethylase, there is significant enhancement in number of H-bonds. Compounds 1, 4 and 8 formed a H-bond whereas propiconazole did not showed any H-bond (Figs. 3, 5, 7, 8). In both the compounds, O-atom directly attached to phenyl spacer and N-atom of benzimidazole/1,2,4-triazole i.e hetero atoms (O and N) played significant role in H-bond formation causing strong binding to enzymes and hence greater docking scores.
Overall, only three compounds viz. 1, 4 and 8 were chosen for synthesis following the combined criteria of stronger binding to tubulin and lanosterol 14α-demethylase (with docking score ≥ −7.3 and −8.9 Kcal/mol, respectively), Lipinski filtrations, FMO approach and Toxtree analysis. In silico guided synthesis of three selected compounds (1 4 and 8) was carried out via multiple steps followed by their antifungal evaluation studies against phytopathogenic fungi of rice.
3.2 Chemistry
The synthesis of novel 1,2,4-triazole linked benzimidazole derivatives (1, 4 and 8) was carried out in three-step reaction process (Scheme 1). The products 1c-1e were synthesized from 1a to 1b with chloroacetyl chloride followed by treatment with corresponding 1,2,4-triazole. The progress of reaction was monitored by IR. The appearance of peaks due to groups, C⚌O (ester), C—N, C⚌N between 1720 and 1730, 1150–1200 and 1300–1500 cm−1, respectively, in addition to aldehydic peaks between 1650 and 1685 (Supplementary material), conformed the formation of 1c-1e. The products 1c-1e were then reacted with o-phenylenediamine in the presence of sodium bisulfite to yield 1, 4 and 8, respectively. All the crude products were recrystallized from ethanol to get pure compounds (conformed by single spot TLC). Formation of 1, 4 and 8 was further confirmed by disappearance of spectral peaks of aldehydic carbonyl group present in 1c-1e and appearance of –NH stretching between 3250 and 3350 cm−1. For example, formation of compound 1 was confirmed by disappearance of peak at 1683 nm in its IR spectrum which was present in that of product 1c. The spectral data (IR, 1H NMR, 13C NMR and mass) of all the synthesized compounds were correlated with the proposed structure which confirmed the formation of the compounds.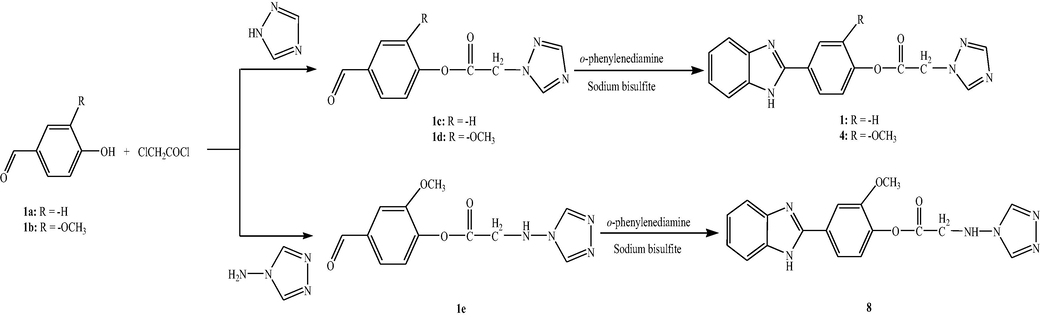
Synthetic route to benzimidazolyl-1,2,4-triazoles.
3.3 Antifungal evaluation of synthesized compounds
The synthesized benzimidazolyl-1,2,4-triazoles (1, 4 and 8) were screened for their in vitro antifungal assay against F. verticillioides, D. oryzae, C. lunata and F. fujikuroi using poisoned food technique and the results were expressed in terms of ED50 values (Table 4). All the three compounds 1, 4 and 8 effectively inhibited the growth of all the test fungi with ED50 values ranging from 10 to 50 μg/ml. These were multi fold lower than the standard carbendazim with ED50 values of 230 and 150 μg/ml against F. verticillioides and F. fujikuroi respectively. Compound 8 exhibited ED50 lower than standard propiconazole against all the test fungi, while compound 4 (ED50-18 μg/ml) also inhibited the hyphal growth of C. lunata at concentration lower than propiconazole. Overall, compound 8 was found to be the best antifungal compound (Fig. 10) showing broad spectrum of inhibitory activities against all the test fungi (ED50 ≤ 16 μg/ml).
Compound
F. vertcillioids
D. oryzae
C. lunata
F. fujikuroi
1
35
50
28
45
4
30
25
18
30
8
16
12
10
15
Carbendazim*
230
–
–
150
Propiconazole*
20
25
22
21
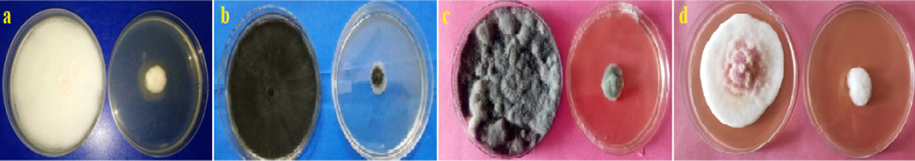
(a–d). Fungal growth of (a) F. vertcillioids, (b) C. lunata, (c) D. oryzae and (d) F. fujikuroi at 20 μg/ml of compound 8.
The results of antifungal evaluation studies were in good agreement with molecular docking studies. Compound 8, with highest docking score in β-tubulin worked well in wet lab studies. LogP also played important role in the inhibitory activity. The logP value of compound 8 was the lowest among the three selected compounds proving higher lipophilicity to the molecules that the other three compounds. Though the docking score of compounds 1 and 4 were in close proximity in both the enzymes, still a visible difference can be attributed to the difference in their logP values. Overall, the highest docking score, lowest logP along with other favourable Lipinski molecular descriptors contributed to the appreciable antifungal potential of compound 8 in comparison to commercial standards used.
3.4 SEM analysis of test fungi treated with compound 8
The detrimental effects of most potent antifungal compound 8 (100 μg/ml) on structural morphology of the all the test fungi was clear form ultra microscopic details. In case of F. verticillioides excessive wrinkling of hyphae and spores along with aberrant topological features were observed. Formation of pits led to the roughness of hyphal cell wall in contrast to smooth surface in control (Fig. 11a-b). The leakage of cellular components from the pits probably led to shrinkage and ultimate cell death. Our observation is in agreement with Nasrollahi et al., (2011) who reported the same effects due to Ag-NPs at a concentration of 4000 μg/ml creating pits in morphology of S. cerevisiae. Compound 8 inhibited the growth of D. oryzae by causing less sporulation and several morphological deformities leading to collapsing of hyphae (Fig. 11d) and severe shrinkage of spores (Fig. 11e). Hyphal surface was quite rough and unsymmetrical as compared to control (Fig. 11c).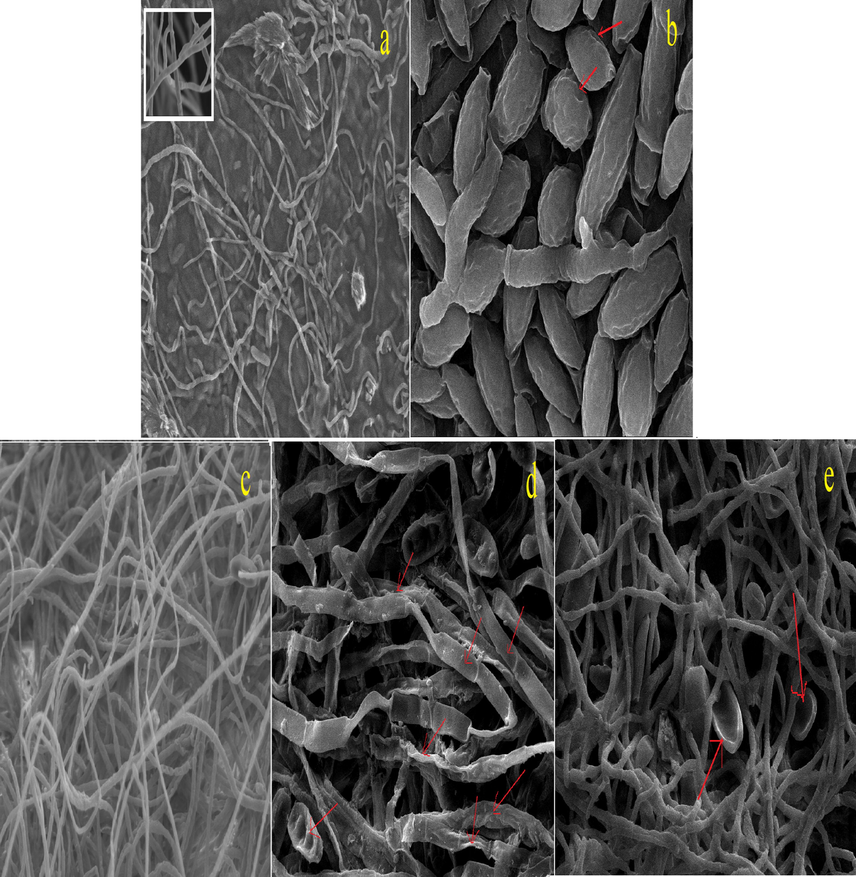
(a–e). Scanning electron microscopy of (a-b) F. verticillioides (a) control (inset-smooth cell wall of hyphae), (b) wrinkling and pits of spore, (c–e) D. oryzae, (c) control (d) deformation and collapsing of hyphae, and (e) collapsing of spores when treated with compound 8 (100 μg/ml).
SEM pictures of untreated C. lunata showed typical cell morphology with normal spores. Shrinkage of spores was observed which reduced the spore size to about 7.23 μm whereas average spore size was 8.3 μm in control (Fig. 12(a and b)). Severe distortion and disruption of fungal hyphae was seen in the pictures (Fig. 12c). Lamsal et al. (2018) reported that spore shrinkage and hyphal distortion of powdery mildews occurred upon treatment with AgNPs. Similar deformities were observed in F. fujikuroi SEM examination confirmed the fungi cell membrane rupture resulting in possible inhibition of fungal growth due to antifungal treatment.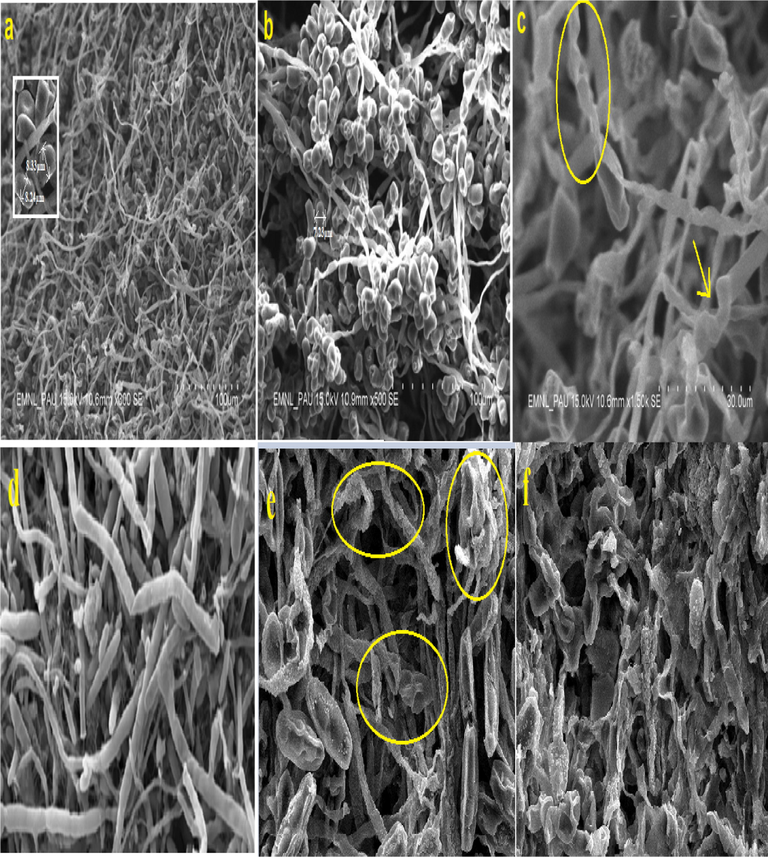
(a–f). Scanning electron microscopy of (a–c) C. lunata (a) control, (b) shrinkage and reduction of spore when treated with compound 8 (100 μg/ml), (c) deformation and disruption of hyphae, (d–f) F. fujikuroi (d) control with smooth cell wall, (e) shrunken spores and deformation of hyphae and (f) severe deformation of hyphae when treated with compound 8 (100 μg/ml).
4 Conclusion
4-(1H-benzo[d]imidazol-2-yl)-2-methoxyphenyl2-(1H-1,2,4-triazol-1-ylamino)acetate (8) demonstrated the best antifungal evaluation results hypothetically twin targeted against β-tubulin and lanosterol 14α-demethylase. The results reinforced the synergistic effects of benzimidazole and 1,2,4-triazole combination supported by computational approach. It is worthwhile to access the combination and to take compound 8 as lead molecule for further research and exploration to develop new antifungal agents with which may help to combat drug resistance with great safety profile.
Declaration of Competing Interest
The authors declared that there is no conflict of interest.
References
- Yeast cytochrome P450 catalyzing lanosterol 14α-demethylation. J. Biol. Chem.. 1984;259:1661-1666.
- [Google Scholar]
- Design, synthesis and biological evaluation of novel benzimidazole amidines as potent multi-target inhibitors for the treatment of non-small cell lung cancer. Eur. J. Med. Chem.. 2018;143:1616-1634.
- [CrossRef] [Google Scholar]
- Fusarium, laboratory guide to the identification of the major species. Mycologia. 1977;69:1239.
- [CrossRef] [Google Scholar]
- Synthesis, molecular docking studies, and antifungal activity evaluation of new benzimidazole-triazoles as potential lanosterol 14α -demethylase inhibitors. J. Chem.. 2017;2017
- [CrossRef] [Google Scholar]
- Synthesis, antimicrobialand molecular docking studies of novel benzimidazole derivatives. IOSR J. Appl. Chem.. 2015;8:28-33.
- [CrossRef] [Google Scholar]
- Current status and future challenges in molecular design for reduced hazard. ACS Sustain. Chem. Eng.. 2016;4:5900-5906.
- [Google Scholar]
- New PCR assays for the identification of Fusarium verticillioides, Fusarium subglutinans, and other species of the Gibberella fujikuroi complex. Int. J. Mol. Sci.. 2011;13:115-132.
- [CrossRef] [Google Scholar]
- In silico design and molecular docking studies of novel pyridyl triazole derivatives as CYP-51 inhibitors. Int. J. Innov. Pharm. Sci. Res.. 2014;2(12):3032-3039.
- [Google Scholar]
- Molecular identification of fusarium species in Gibberella fujikuroi species complex from rice sugarcane, and maize from Peninsular Malaysia. Int. J. Mol. Struct. 2011:6722-6732.
- [CrossRef] [Google Scholar]
- International Rules for Seed Testing. Zurich, Switzerland: ISTA; 1993. p. :345.
- Design, synthesis and antifungal evaluation of novel benzimidazole tertiary amine type of fluconazole analogues. Arab. J. Chem.. 2019;12:2224-2235.
- [CrossRef] [Google Scholar]
- Molecular genetic analysis of azole antifungal mode of action. Biochem. Soc. Trans.. 1993;21:1034-1038.
- [CrossRef] [Google Scholar]
- Identifying and designing chemicals with minimal acute aquatic toxicity. P. A. N. S.. 2014;112:6289-6294.
- [CrossRef] [Google Scholar]
- Inhibition effects of silver nanoparticles against powdery mildews on cucumber and pumpkin inhibition effects of silver nanoparticles against powdery mildews on cucumber. Mycobilogy 2018:8093.
- [CrossRef] [Google Scholar]
- LigPlot: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model.. 2011;51:2778-2786.
- [CrossRef] [Google Scholar]
- Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev.. 2012;64:4-17.
- [CrossRef] [Google Scholar]
- ACS Med. Chem. Lett.. 2014;5:846-850.
- A species-specific PCR assay based on the calmodulin partial gene for identification of Fusarium verticillioides, F. proliferatum and F. subglutinans. Eur. J. Plant Pathol.. 2004;110:495-502.
- [CrossRef] [Google Scholar]
- Antifungal activity of silver nanoparticles on some of fungi. Rev. Inst. Med. Trop. Sao Paulo. 2011;1:233-239.
- [Google Scholar]
- Nene, N.L., Thapliyal, P.N., 1997. Fungicides in Plant Disease Control, New Delhi, India.
- Comparative analysis of molecular and biological characteristics of strains of Beauveria brongniartii isolated from insects. Mycol. Res.. 1994;98:322-328.
- [CrossRef] [Google Scholar]
- Design, synthesis, and antifungal activity of triazole and benzotriazole derivatives. Eur. J. Med. Chem.. 2009;44:3064-3067.
- [Google Scholar]
- Synthesis of novel fluorinated benzothiazol- 2-yl-1,2,4-triazoles: Molecular docking, antifungal evaluation and in silico evaluation for SAR. Arab. J. Chem. 2015
- [CrossRef] [Google Scholar]
- Disease Note: Identification of Curvularia oryzae as cause of leaf spot disease on oil palm seedlings in nurseries of Thailand. Phytoparasitica. 2014;42:529-533.
- [CrossRef] [Google Scholar]
- Selecting the right compounds for screening: does Lipinskis Rule of 5 for pharmaceuticals apply to agrochemicals? Pest Manag. Sci.. 2001;57:3-16.
- [CrossRef] [Google Scholar]
- AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Chem. Inf. Model. 2009:455-461.
- [CrossRef] [Google Scholar]
- Some principles of fungicide resistance. Plant Pathol. Fact Sheet. 2014;1(1):123-125.
- [Google Scholar]
- Design, synthesis and evaluation of clinafloxacin triazole hybrids as a new type of antibacterial and antifungal agents. Bioorg. Med. Chem. Lett.. 2012;22:5363-5366.
- [CrossRef] [Google Scholar]
- Genetic variation among pathogens causing “Helminthosporium” diseases of rice, maize and wheat. FitopatologiaBrasileira. 2002;27:639-643.
- [CrossRef] [Google Scholar]
- J. Chem. Inf. Comput. Sci.. 1988;28:31-36.
- [CrossRef]
- Towards rational molecular design: derivation of property guidelines for reduced acute aquatic toxicity. Green Chem.. 2011;13:2373.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2020.04.020.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary Data 1
Supplementary Data 1







