Translate this page into:
Untargeted screening of plant metabolites based on data-independent and data-dependent acquisition modes using LC-ESI-QTOF-MS: Tribulus terrestris L. as a case study
⁎Corresponding author at: H.E.J. Research Institute of Chemistry, International Center for Chemical and Biological Sciences, University of Karachi, Karachi 75270, Pakistan. musharraf@iccs.edu (Syed Ghulam Musharraf)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
Metabolomics has been used as a powerful tool for the analysis, and drug-lead identification in medicinal plants and herbal medicines. For the coverage of a broader range of plant-based metabolites using LC-MS, one of the important parameters is the selection of analysis mode and data processing for identification. This paper describes the utility of two distinctive acquisition modes in combination, a classic data-dependent acquisition (DDA) mode and a data-independent acquisition (DIA) mode for obtaining the mass spectrometry data of plant extracts using LC-ESI-QTOF/MS. Tribulus terrestris plant was used as a case study. We have applied three-step data analysis pipeline 1-annotation and putative identification of metabolites, 2-validation, and relative quantification, and 3-multivariate analysis using open-access MS-DIAL, Skyline, and Perseus software. A total of four samples of T. terrestris (aqueous extracts), two fruits, and two whole plant samples, from two different regions, were used. By combining data analysis results of plant fruit samples from two different regions, a total of 95 and 77 metabolites were identified in positive and negative ionization modes, respectively. Similarly, in the analysis of the whole plant from two different regions, 75 and 76 metabolites were identified in positive and negative ionization modes, respectively. We suggested the use of DDA mode for annotation, identification of metabolites, and generation of transition lists in MS-DIAL, furthermore, the use of DIA acquisition mode for enhancing metabolites sensitivity in complex samples, deconvolute MS1/MS2 spectra in Skyline for the quantitative performance and analytical reliability. The developed protocol can be used for the broader coverage of plant-based metabolites.
Keywords
Data-independent acquisition (DIA)
MS-DIAL
Skyline
Tribulus terrestris
Liquid-chromatography-mass-spectrometry (LC-MS)
1 Introduction
Metabolomics has grown into one of the major approaches for drug discovery studies, in part driven by developments in mass spectrometry (MS), providing high sensitivity and high-throughput coverage of the metabolome (Wolfender et al., 2009). Untargeted metabolomics allows for the investigation of metabolic phenotypes involving several hundred to thousands of metabolites. Moreover, metabolomics is a powerful platform for the comprehensive profiling of a series of small-molecule metabolites (MV < 1000). Currently, the most promising technique in terms of metabolome coverage is liquid chromatography coupled to mass spectrometry (LC-MS; Steiner et al., 2021; Pokrywka et al., 2014). Compared to other alternatives, LC-MS is more versatile in that sample preparation and elution systems (chromatographic separation) can be adapted to nearly every class of compound while being extremely sensitive and additionally providing rich structural information (Pokrywka et al., 2014). These features make LC-MS a particularly interesting choice for untargeted metabolomics, i.e., the unbiased measurement of all metabolites detected in a particular analysis.
The untargeted metabolomics experiment aims to process every signal and try to identify relevant signals by combining two LC-MS acquisitions into larger datasets that can reach up to several thousand signals (Perez de Souza et al., 2019). The mass spectrometry data were respectively obtained by two typical acquisition modes, data-dependent acquisition (DDA) and data-independent acquisition (DIA) modes. A recent trend that has been developing in the last few years and is particularly useful in untargeted metabolomics is the data-independent acquisition (DIA) of tandem mass spectra (Guo and Huan 2020). Commonly used DIA methods include all-ion fragmentation (AIF) (e.g., MSAll. MSE), where all precursors are fragmented, and sequential window acquisition of all theoretical fragment-ion spectra (SWATH) where a medium pass window, such as 20 or 25 Da, is used. In traditional data-dependent acquisition, specific ions were selected and fragmented providing a link between parental ions and daughter fragments. However, DIA uses much broader windows for the selection of parental ions, resulting in a second-order spectrum containing fragments from all ions in the first-order spectra, potentially providing richer structural information but with a disadvantage: loss of the parental/daughter ion relationship (Perez de Souza et al., 2019; Guo and Huan 2020). In previous studies, some research groups have developed software tools focused on the processing of DIA-MS data to facilitate the annotation and identification process by deconvoluting MS2 spectra(Perez de Souza et al., 2019, Guo and Huan 2020).
Tribulus terrestris L. a crawling plant belonging to the family, Zygophyllaceae. It is widely occurring in sub-continents i.e., Africa, Australia, Asia, and Europe. The name of the plant is coined from the Greek word “tribolos” meaning spike fruit (Ma et al., 2017). It is used to re-cover visual sharpness and epilepsy in Traditional Chinese Medicine (TCM) while used for its applications as diuretic, stimulant, and aphrodisiac properties in Indian herbal medicine. The TCM name of T. terrestris fruit is “Ji Li” which holds for several activities, such as activities against heart diseases (Neychev and Mitev 2016), eye inflammation, skin irritation (Reshma et al., 2016), and abdominal distention. The main use of the plant in Europe and the USA is for the increase in muscle strength and improvement of sexual function (Saleem et al., 2020; Angelova et al., 2013), whereas in China, it is used for the treatment of cardiovascular diseases (Ștefănescu et al., 2020). The medicinal importance of T. terrestris, indicated against neurodegeneration is due to the presence of steroidal saponins, alkaloids, flavonoids, steroidal glycosides, cinnamic acid, and phytosterol (El-Shaibany et al., 2015).
T. terrestris extracts have been tested for various effects like cardioprotective, hepato-protective, antitumor (Pokrywka et al., 2017; Figueiredo et al., 2021), anti-atherosclerotic, anti-arthritic (Li et al., 2013; Mishra et al., 2013), antioxidant and antimicrobial (Sivapalan 2016; Akbal et al., 2018), analgesic and anti-inflammatory properties, and could be used for the ailment of oral infections and control of dental problems (Oh et al., 2012). It has been studied for its effectiveness in the in-vivo treatment of hyperglycaemia and in-vitro inhibition of α-glucosidase, aldose reductase, and in the treatment of pancreatitis in mice (Soleimanpour et al., 2015; Lamba et al., 2011). In a previously reported study, phytochemicals of T. terrestris have been investigated (Zheng et al., 2017).
In this regard, we present herein a comprehensive untargeted metabolomics study of T. terrestris plant for the putative identification of wide range metabolome and relative quantification by using combinations of DDA and DIA modes using LC-ESI-QTOF/MS. This is the first broad strategy for untargeted screening of phytochemicals in complex samples by combining results of DDA and DIA modes (to the best of our knowledge). Various studies reported a comparison of untargeted metabolomics results between DDA and DIA mode, but not for untargeted phytochemical analysis (Barbier Saint Hilaire et al., 2020, Guo and Huan 2020, Liu et al., 2021). A previous study highlighted the specific class of phytochemicals using the DIA acquisition mode (Liu et al., 2021). The study provides an efficient workflow for untargeted plant-based metabolomics, which can be useful for the rapid dereplication of plant extracts and for monitoring the quality control of herbal medicines.
2 Materials and methods
2.1 Chemicals and reagents
All solvents were purchased commercially. Methanol (Analytical grade) solvent was purchased from Merck K GaA, 64,271 (Darmstadt, Germany). Formic acid was purchased from Daejung (Daejung chemicals and Metals Co. Ltd, Korea). In house, a water purification system (Thermo Scientific, USA), was utilized to get high-purity water (resistivity 18.1 MΩ cm at 25 °C).
2.2 Sample preparation
T. terrestris plant samples were collected from two different regions of Pakistan including Peshawar (Region 1) and Karachi (Region 2) and were identified by the plant taxonomist. Shade-dried plant material (fruit and whole plant) were crushed in a blender separately. 1 g of each plant powder was weighed and extracted with a 10 mL mixture of methanol and water (8:2) through sonication for 20 min. Each sample was centrifuged for 15 min at 6000 rpm to settle large particles, and the supernatant was filtered through a 0.22 µm PTFE syringe-driven filter. 50 µL of the filtered extract was diluted to 1000 µL with methanol for LC-MS, and LC-MS/MS analysis.
2.3 Instrumentation and experimental conditions
All acquisitions were performed using Bruker maXis II HR-QTOF mass spectrometer (Bremen, Germany), coupled with the Dionex Ultimate 3000 series UPLC system (Thermo Fischer Scientific Waltham, MA, USA), further facilitated with auto-sampler, column thermostat, and binary RS pump. For the stationary phase, the Macherey-Nagel™ Gravity column (3.0 × 100 mm, 1.8 μm), was used. The mobile phase consisted of 0.1% formic acid in water (A), as well as in methanol (B). Linear gradient solvent composition was used for HPLC starting with 10% B and raised to 90% B in 6.5 min, which was maintained at 90% B for 1.5 min and then decreased back to 10% B in 1.0 min, 1 min equilibration time was given before and after the gradient. Furthermore, in positive and negative ion modes, 5 μL of the samples were injected into LCMS. A capillary voltage of 4500 V; end plate offset of 500 V; drying gas (nitrogen) with a flow rate of 10 mL/min; drying gas temperature of 280 °C; and nebulizer gas pressure of 45.0 psi, were optimized for this study. For data-dependent acquisitions (DDA), auto MS/MS mode was used with the mass range of 70–1200 m/z, isolation window typically < 2 Da, exclusion time of precursor ion 5 s; a scan speed of 20 Hz, and a range of collision energies (CE); 10, 20, 30, 40, and 60 eV. For data-independent acquisitions (DIA), Q1 window of 25 and 50 Da was used within the mass range of 99–1200 m/z (Table S-1) with the collision energy of 30 eV (the only CE was best fitted in DDA acquisition).
2.4 Software and data processing
The DDA data acquired for samples in raw data (.d) format was first converted into the Analysis Base File (Abf.) format. MS-DIAL ver 4.80 software (http://prime.psc.riken.jp/compms/msdial/main.html) was used for feature detection, ion species annotation, compound spectra extraction, and peak alignment between samples and spectral analysis resulting in the transition list containing information on every detected feature such as retention time, precursor m/z, and MS2 ions. Identification of untargeted metabolites in the complex mixture has been done in MS-DIAL using MS/MS libraries available (MS/MS-Pos and MS/MS-Neg). The MS-DIAL parameters were set as follows: retention time 1–13 min; mass range 70–1200 Da; minimum peak width, 5 scans; minimum peak height, 1000 amplitude; smoothing level, 2; MS/MS abundance cut off, 10 amplitude; mass slice width, 0.1 Da; exclusion mass list, none; retention time tolerance, 2 min; MS1 accurate mass tolerance, 0.01 Da; MSMS accurate mass tolerance, 0.05 Da; identification score cut off, 70%. Spectral library, (MS/MS-pos and neg) LC-MS/MS in MSP format.
DIA acquisition mode was used to “Dig deep” for every identified metabolite in multiple DDA run for every sample. Skyline (21.1.0.146) is open-source software (https://skyline.ms/project/home/begin.view) well known in proteomics for its unique features especially visualization and advanced method refinement options such as retention time scheduling and reproducibility (Adams et al., 2020). These features are now also implemented for DIA workflows of small molecules such as pharmaceuticals (drug metabolites, and herbal medicines), metabolomics, forensics, and food safety (Rardin 2018). The Skyline parameters for DIA were set as, TOF for precursor mass analyzer, resolving power; 30,000, mass range; 70–1200 Da, retention time filtering. An isolation window with a gap of 50 and 25 Da. covering a mass range of 99.5–1200 Da. for MS2 data. The method has a margin of 0.5 m/z added to the isolation window, for example, 99.5–150.5, 149.5–200.5 etc.
2.5 Multivariate analysis
A.CSV file containing peak areas of each sample was imported by using generic matrix upload in Perseus (https://maxquant.net/perseus/) and the data were transformed to a logarithmic scale (log2(x)). To visualize the sample correlations, the “Analysis Hierarchical clustering” option was used for clustering all replicates of samples based on the correlation coefficients between them revealing higher similarity on MS1 levels. The principal component analysis (PCA) and loading plots (Orthogonal Projections to Latent Structures Discriminant Analysis, OPLS-DA) were plotted using SIMCA (Version 15.0, Umertrics Umeå, Sweden) software (https://www.sartorius.com/en/products/process-analytical-technology/data-analytics-software/mvda-software/simca).
3 Results
This study suggested a pipeline of mass spectrometry acquisition modes (data-dependent and data-independent acquisition) and bioinformatics tools for complex samples like plant extracts. Here, we present an effective workflow based on open-access software for comprehensive untargeted analysis including feature generation, putative identification, relative quantification, and multivariate analysis using MS-DIAL, Skyline, and Perseus software. We used the whole plant, and fruit extracts of T. terrestris to establish a pipeline of mass spectrometry-based data analysis.
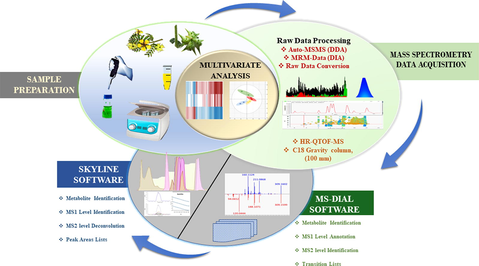
In this study, we proposed a three-step strategy, the first step involved in feature generation, annotation and untargeted identification of metabolites, followed by transition list generation of plant metabolites in each sample using DDA mode in MS-DIAL software. In the second step, a comprehensive transition list containing precursor ions, product ions, and retention time of each feature was imported in Skyline software for processing DIA mode and extracted MS1/MS2 ion chromatograms for deconvolution and relative quantification. The manual integration where required and the usual inspection for features list on MS1 and MS2 levels were inspected. In the third step, the peak areas of metabolites were exported to Perseus software for multivariate analysis. The schematic illustration showed the pipeline of the proposed methodology in Fig. 1.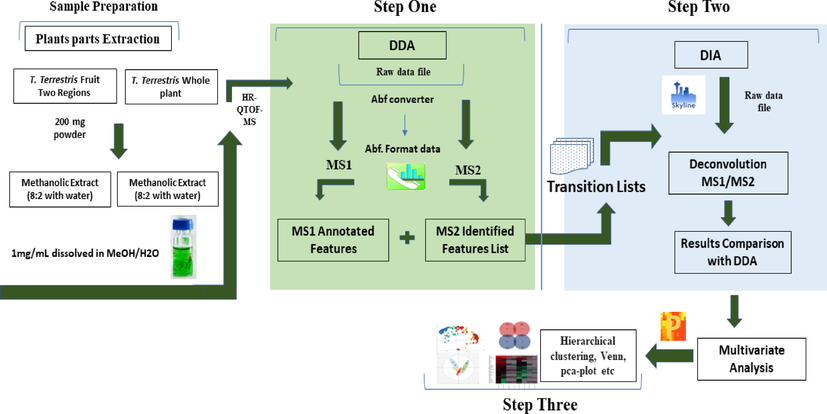
Schematic illustration of the workflow used in this study.
3.1 Identification by DDA in MS-DIAL
In positive ion mode, 362 and 329 MS1 features were annotated out of 1396 and 1240 (remaining are unknown features or not under defined ppm tolerance) in the fruit samples from region 1 and region 2, respectively. From annotated metabolites, 51 metabolites in region 1 and 37 metabolites in region 2 were identified by MS2 respectively. Similarly, 312 and 323 MS1 features were annotated out of 1570 and 1445 in the whole plant extract from both region 1 and region 2, respectively. 49 metabolites in region 1 sample and 52 metabolites in region 2 sample were identified by MS2 data. In negative ion mode, 610 MS1 features were annotated, and 78 metabolites were identified by MS2 data by combining the results of both region samples. All metabolites identification was optimized under 10 ppm mass error and >300 dot formula. (Table S-2, 3). The dot product score was used in combination with retention time and accurate mass, to increase the confidence in the identification of metabolites. The exact threshold for the dot product score may vary depending on the specific dataset and the quality of the reference library used. The peak areas of each identified feature in each sample were used for multivariate analysis.
3.2 Deconvolution of MS2 spectra by DIA in Skyline
A raw transition list from DDA data containing information on precursor and fragments ions of metabolites generated from MS-DIAL was used for achieving better spectral quality and quantity of MS2 deconvolution in DIA using Skyline. DIA mode is described in this method to achieve sensitivity, resolve co-elution, and relative quantification by peak areas. An isolation window of 25 and 50 Da was used for automatically extracting MS1 and MS2 ion chromatograms from transition lists. Skyline provides easy observation over retention time, extracted ion chromatograms, and peak areas for each metabolite across numerous samples or replicates. In positive ion mode, 95 and 75 metabolites were found in DIA fruit samples from region 1 and region 2, with 48 and 33 unique metabolites in both regions, respectively. Similarly, 75 and 75 metabolites were found in whole plant samples from region 1 and region 2, with 26 and 23 unique metabolites in both regions, respectively. While, in negative ion mode, 78 metabolites were found in DIA by combining both region sample results under 10 ppm mass error. All identified metabolites in positive ion mode from region 1 and region 2 comparison results are illustrated in the Venn diagram Fig. 2.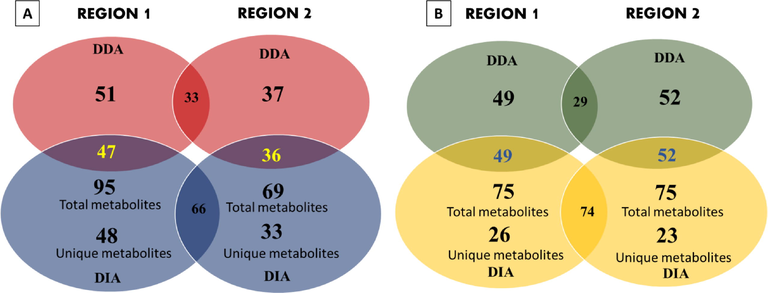
Metabolites distribution difference in T. terrestris fruit and whole plant samples from two different regions. (A) Venn diagram of T. terrestris fruit metabolites (B) Venn diagram of T. terrestris whole plant metabolites.
Unique Skyline visualizations are helpful for data interrogation and useful to easily visualize the peak intensities of the targeted metabolite among the samples. As an example, in the Fig. 3A, DIA mode was able to find a metabolite (tranilast m/z: 326.1039) by MS2 spectra despite missed MS1 peak. Additionally, the deconvolution of MS2 data for metabolite (spirostane + 1O m/z: 929.4760) in DIA provides an advantage for coeluting isobaric and high background metabolites by using specific transitions for relative quantitative analysis (Fig. 3B). DDA is generally employed with exclusion criteria that allow deeper MS2 data for low abundant metabolites. However, MS2 data cannot be used for the quantitative purpose. Here, DIA is superior as it can be used for the quantification of both MS1 and MS2 levels. The peak areas of metabolite (flavonol m/z: 493.1018) at both levels (MS1/MS2) can easily be extracted from Skyline to use for multivariate analysis (Fig. 3C).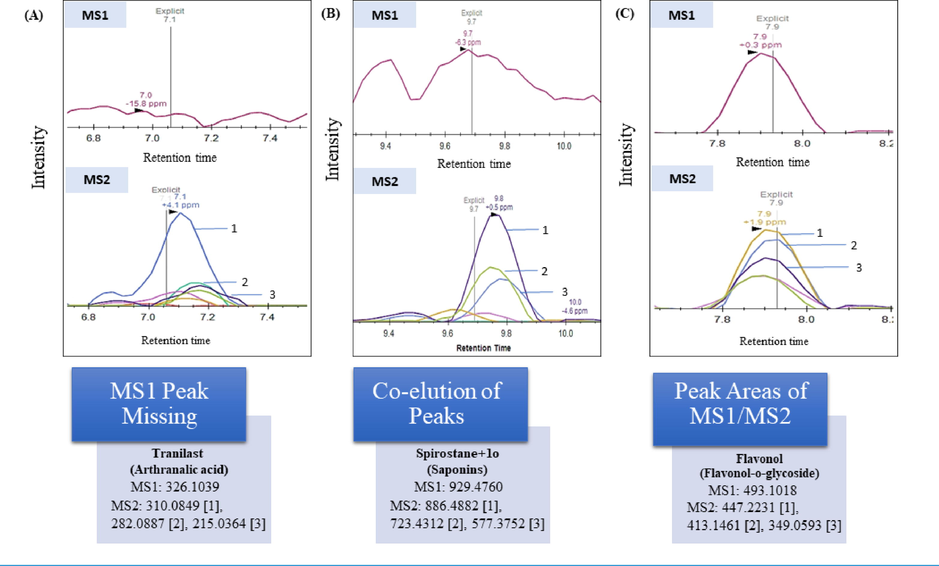
Applications of DIA acquisition mode. (A) The confirmation of metabolite by MS2 spectra despite missed MS1 peak. (B) The advantage of using Skyline for DIA MS2 deconvolution is to minimize the factor of co-elution of MS1 peaks in complex samples. (C) Skyline provides a clear illustration of DIA MS1 and MS2 spectra for quantitative studies.
3.3 Metabolite clustering
Based on the ion peak areas observed, heat maps were generated for both ionization modes (positive and negative) to show a distribution of various plant metabolites across two regions. The detection and hierarchical clustering of all identified metabolites in plant fruit and ariel part samples with their MS1 datasets represent peak areas in each sample shown in Fig. 4. Hierarchical clustering of metabolites in negative ionization mode is presented in supplementary file Fig. S1. The content of metabolites in T. terrestris was found lower in the Karachi region compared to the content of the Peshawar region. Results obtained in DDA mode, the distribution of metabolites in both regions was found different. The heatmaps analysis showed that the highest concentrations of compounds having several biological activities including, flavonoids, alkaloids, terpenoids, and saponins are present in the sample of the Peshawar region. Results obtained from the DIA mode proved the wide range of metabolite analysis in samples of both regions and eventually observed clear difference in the distribution of metabolites between both samples.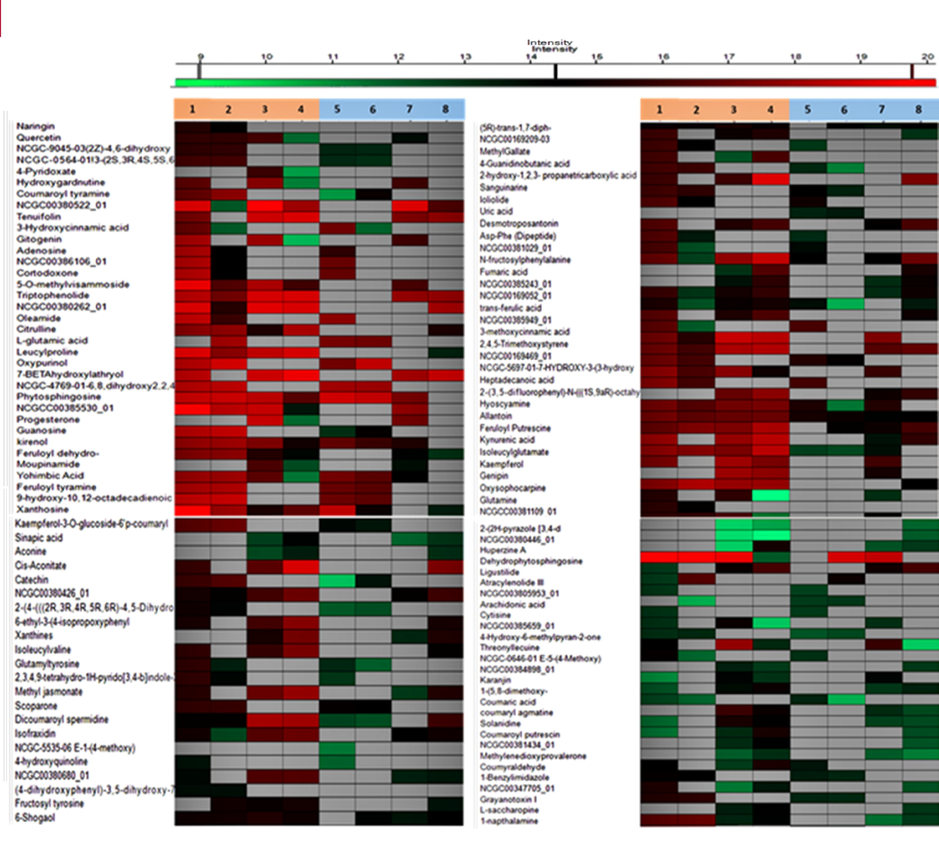
Hierarchical clustering of total of 118 metabolites identified (by using MS1 data) in each sample of plant (F* = fruit, WP* = Whole plant, R = Region) in pos. ion mode were generated using Perseus software. 1. DIA F* R1, 2. DIA F* R2, 3. DIA WP* R1, 4. DIA WP* R2, 5. DDA F* R1, 6. DDA F* R2, 7. DDA WP* R1, 8. DDA WP* R2.
The PCA and OPLS-DA plots illustrated valuable information that is helpful for the understanding of the variation of phytochemicals between the groups. After Pareto scaling and mean centering, PCA score scatter plot was plotted, results showed a distribution and variation within and among the samples as shown in Fig. 5. A separation was observed among the eight groups whereas, the fruit DIA sample has shown larger variation within the groups as compared to the other groups.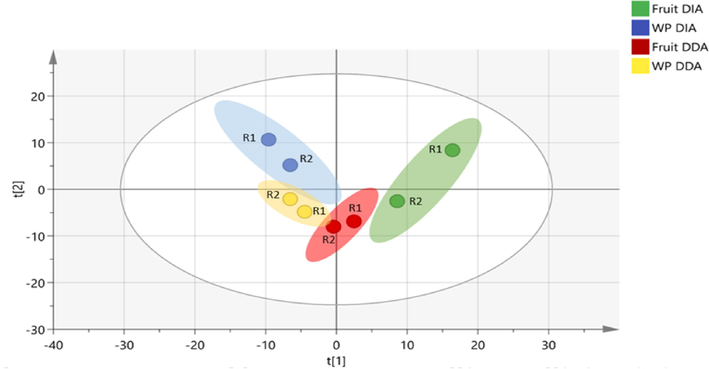
PCA scores scatter plot of 8 groups in which each colour represents a sample’s type and each dot represents a sample having confidence limit of Hotelling’s T2 test is 95%.
Moreover, a loading plot was also generated based on p-values and coded all features according to their p-values as shown in Fig. 6. A total of 18 features are highly influential in the discrimination of the groups. These 18 features were found in each sample with different peak areas and intensities.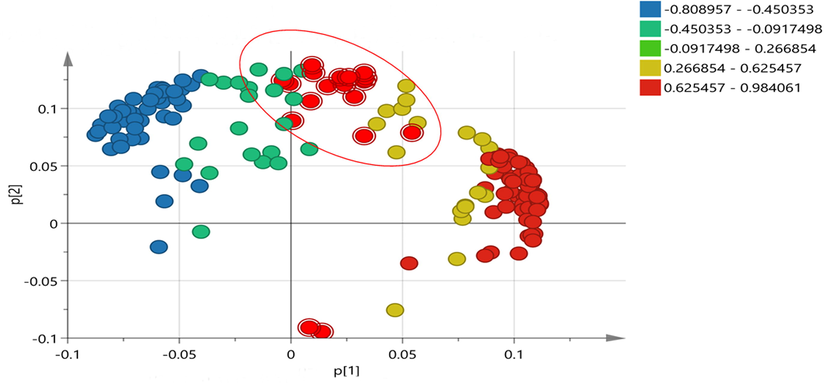
OPLS-DA loading plot of all features extracted from the mass spectra. The red features in the plot showed the lowest p-values, indicates that these are highly involved in dis-crimination of the groups.
4 Discussion
The difference in results observed in both acquisition modes is due to the biased selection of precursor ions in the DDA mode. The MS2 spectra are automatically collected for pre-cursor ions whose MS1 intensity matched or exceeded a pre-defined threshold value. In DDA acquisition mode, precursor ions are selected using a small isolation window (typically ≤ 2 Da wide), which results in high-quality and pure MS2 spectra. The loss of many precursor ions in DDA analysis is due to these ions are not selected for fragmentation if they do not match the defined threshold value (low intense peaks) or if they are co-eluted with other intense peaks. However, in the DIA acquisition, the precursor ions are selected using a wide m/z isolation window (typically between 25 and 50 Da) to cover the whole mass range. All ions within these windows are then fragmented unbiasedly and simultaneously. Therefore, both modes are highly complementary and each of them has been utilized to bring qualitative and quantitative information to this study. Furthermore, the difference between the sensitivity of both methods by their chromatograms in plant fruit samples is highlighted in Fig. 7. The base peak chromatograms illustrate the difference between sensitivity in both methods very sharply. The DIA method has achieved higher sensitivity and generated better qualitative profiling of phytochemicals in complex samples. The results showed many low intense peaks were detected in DIA acquisition which were missed in the DDA fruit sample due to low intensity or eluted below by given threshold value. As we see in Fig. 5, the compound 1 C11H20N2O3, m/z: 229.1544 (M + H)+, compound 2 C22H22O5, m/z: 367.1495 (M + H)+, and compound 3 C32H38O19, m/z: 727.2060 (M + H) + have achieved better sensitivity; moreover, with enhanced sensitivity DIA mode was able to dig deeper to detect low intense metabolites in the complex plant sample. DIA mode achieved a sensitive and highly qualitative profile of each plant sample from both regions.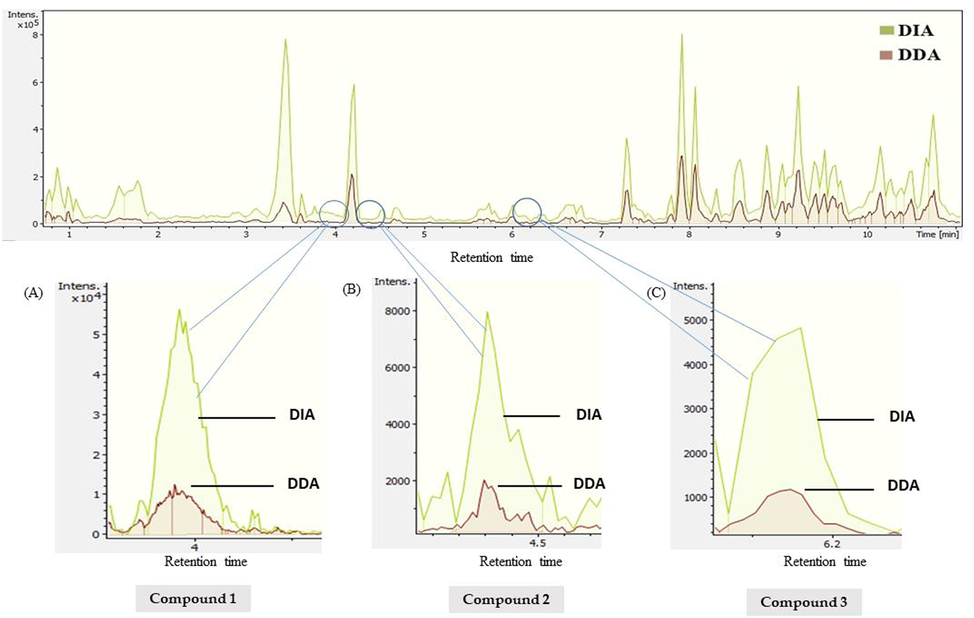
Comparison of method sensitivity in DIA and DDA modes by base peak chromatograms (BPCs) in plant fruit sample. (A) C11H20N2O3, m/z. 229.1544 (M + H)+; (B) C22H22O5, m/z. 367.1495 (M + H)+; (C) C32H38O19, m/z. 727.2060 (M + H) +.
Hierarchical clustering of all metabolites in each sample was generated using Perseus software. As Perseus is designed for proteomics data, unlike other commercial software it is optimized to use at large numbers of variables and is a very powerful software for multivariate analysis of high dimensional data, thus is well suited for metabolomics data also (Tyanova and Cox 2018).
We suggest the use of MS-DIAL software for feature generation, annotation, identification, and MS2 database search followed by a comprehensive transition list generation using DDA mode in MS-DIAL. This transition list can be uploaded to Skyline for dig-deep strategy and relative quantification in DIA mode. Perseus software for multivariate analysis of metabolites in a complex sample using the efficiency of DDA and DIA mode.
Moreover, the developed strategy may be useful for bioactivity-guided lead molecule discovery in natural products and can be achieved through bioassays of various fractions of plant extracts and then the activity of a fraction can be attributed to the presence of specific natural products in that fraction. T. terrestris contains high content of active ingredients (in particular sterol saponins (Semerdjieva and Zheljazkov, 2019), as well as flavonoids, tannins, terpenoids, phenol carboxylic acids, and alkaloids), and its frequent uses in folk medicine and numerous herbal supplements are commercialized worldwide with indications mostly to improve libido, sexual performance in both sexes, and athletic performance, common cough, viral flu, cold, asthma and inflammatory diseases [6–10]. The pharmacological effect of the plant with different diseases in metabolite-diseases interaction network has already been published and discussed in recent literature (Ștefănescu et al., 2020, Amanullah et al., 2021).
5 Conclusion
In conclusion, we have successfully utilized a combination of DDA and DIA-based mass spectrometry strategy for the identification of a wide range of metabolites in the medicinal and industrial valued plant, T. terrestris (as a case study). Moreover, a pipeline that integrates freely available software, MS-DIAL, Skyline, and Perseus for the metabolomic profiling was employed. This strategy will be helpful to overcome the limitation with DDA in that the precursors with low MS abundance will not get fragmented in a single experiment due to low intensity. We suggested the implementation of DIA mode for the MS2 deconvolution and relative quantification of untargeted metabolites in the field of plant-based metabolomics, specifically for ones with less abundance. This approach can be used as a method of choice for the large-scale profiling and rapid analysis of the metabolites in medicinal plants as well as a quality control tool for herbal formulations.
Funding
The authors (Mufarreh Asmari & Jalal Uddin) would like to extend their appreciation to the Deanship of Scientific Research at King Khalid University for supporting this work through the small research group grant number RGP.1/312/44.
Acknowledgement
Syed Muhammad Zaki Shah would like to acknowledge the Higher Education Commission of Pakistan's Indigenous Ph.D. Fellowship program and the Commonwealth Scholarship Commission in the UK for assisting with the research.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Skyline for small molecules: a unifying software package for quantitative metabolomics. J. Proteome Res.. 2020;19:1447-1458.
- [Google Scholar]
- Saponin loaded montmorillonite-human serum albumin nanocomposites as drug delivery system in colorectal cancer therapy. Appl. Clay Sci.. 2018;166:214-222.
- [Google Scholar]
- A comprehensive review on a Unani medicinal plant: Tribulus terrestris Linn. Int. J. Herb. Med.. 2021;9:23-28.
- [Google Scholar]
- Antitumor activity of Bulgarian herb Tribulus terrestris L. on human breast cancer cells. J. BioSci. Biotechnol.. 2013;2
- [Google Scholar]
- Comparative evaluation of data dependent and data independent acquisition workflows implemented on an orbitrap fusion for untargeted metabolomics. Metabolites. 2020;10:158.
- [Google Scholar]
- Anti-hyperglycaemic activity of Tribulus terrestris L aerial part extract in glucose-loaded normal rabbits. Trop. J. Pharm. Res.. 2015;14:2263-2268.
- [Google Scholar]
- Antiglycation and antitumoral activity of Tribulus terrestris dry extract. Avicenna J. Phytomed.. 2021;11:224.
- [Google Scholar]
- Comparison of full-scan, data-dependent, and data-independent acquisition modes in liquid chromatography–mass spectrometry based untargeted metabolomics. Anal. Chem.. 2020;92:8072-8080.
- [Google Scholar]
- α-glucosidase and aldose reductase inhibitory activity in vitro and antidiabetic activity in vivo of Tribulus terrestris L. (dunal) Int. J. Pharm. Pharm. Sci.. 2011;3:270-272.
- [Google Scholar]
- Cellular and molecular mechanisms in vascular smooth muscle cells by which total saponin extracted from Tribulus terrestris protects against artherosclerosis. Cell. Physiol. Biochem.. 2013;32:1299-1308.
- [Google Scholar]
- Systematic Characterization and Identification of Saikosaponins in Extracts from Bupleurum marginatum var. stenophyllum Using UPLC-PDA-Q/TOF-MS. Front. Chem.. 2021;9
- [Google Scholar]
- Tribulus terrestris extracts alleviate muscle damage and promote anaerobic performance of trained male boxers and its mechanisms: Roles of androgen, IGF-1, and IGF binding protein-3. J. Sport Health Sci.. 2017;6:474-481.
- [Google Scholar]
- Anti-arthritic activity of Tribulus terrestris studied in Freund's Adjuvant induced arthritic rats. J. Pharm. Educ. Res.. 2013;4:41.
- [Google Scholar]
- Pro-sexual and androgen enhancing effects of Tribulus terrestris L.: fact or fiction. J. Ethnopharmacol.. 2016;179:345-355.
- [Google Scholar]
- Oh, J.S., Baik, S.H., Ahn, E.-K., et al., 2012. Anti-inflammatory activity of Tribulus terrestris in RAW264. 7 Cells (54.2), Am Assoc Immnol.
- Mass spectrometry-based untargeted plant metabolomics. Curr. Protocols Plant Biol.. 2019;4:e20100.
- [Google Scholar]
- Insights into supplements with Tribulus Terrestris used by athletes. J. Hum. Kinet.. 2014;41:99.
- [Google Scholar]
- An Overview on Tribulus terrestris in Sports Nutrition and Energy Regulation. Sustain. Energy Enhanced Hum. Funct. Act. 2017:155-165.
- [Google Scholar]
- Rapid assessment of contaminants and interferences in mass spectrometry data using skyline. J. Am. Soc. Mass Spectrom.. 2018;29:1327-1330.
- [Google Scholar]
- Mitochondrial dysfunction in H9c2 cells during ischemia and amelioration with Tribulus terrestris L. Life Sci.. 2016;152:220-230.
- [Google Scholar]
- Anti-Parkinson’s Activity of Tribulus terrestris via Modulation of AChE, α-Synuclein, TNF-α, and IL-1β. ACS Omega. 2020;5:25216-25227.
- [Google Scholar]
- Chemical constituents, biological properties, and uses of Tribulus terrestris: A review. Natural Prod. Commun.. 2019;14(8):1934578X19868394
- [Google Scholar]
- Biological and pharmacological studies of Tribulus terrestris Linn-A review. Int. J. Multidiscip. Res Dev.. 2016;3:257-265.
- [Google Scholar]
- Antibacterial activity of Tribulus terrestris and its synergistic effect with Capsella bursa-pastoris and Glycyrrhiza glabra against oral pathogens: an in-vitro study. Avicenna J. Phytomed.. 2015;5:210.
- [Google Scholar]
- A comprehensive review of the phytochemical, pharmacological, and toxicological properties of Tribulus terrestris L. Biomolecules. 2020;10:752.
- [Google Scholar]
- Challenges and future directions in LC-MS-based multiclass method development for the quantification of food contaminants. Anal. Bioanal. Chem.. 2021;413:25-34.
- [Google Scholar]
- Perseus: a bioinformatics platform for integrative analysis of proteomics data in cancer research. In: Cancer Systems Biology. Springer; 2018. p. :133-148.
- [Google Scholar]
- MS-based plant metabolomic approaches for biomarker discovery. Natural Prod. Commun.. 2009;4:1934578X0900401019
- [Google Scholar]
- Rapid characterization of constituents in Tribulus terrestris from different habitats by UHPLC/Q-TOF MS. J. Am. Soc. Mass Spectrom.. 2017;28:2302-2318.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary material to this article can be found online at https://doi.org/10.1016/j.arabjc.2023.104978.
Appendix A
Supplementary material
The following are the Supplementary material to this article:Supplementary data 1
Supplementary data 1







