Translate this page into:
Evaluation of in vitro, in silico antidiabetic and antioxidant potential of bioactivity based isolated “Pakistanine” from Berberis baluchistanica
⁎Corresponding authors at: Department of Pharmacy, The Sahara College Narowal, Narowal, Punjab, Pakistan(M U K Sahibzada), Faculty of Pharmacy, Capital University of Science and Technology, Islamabad, Pakistan(Muhammad Tariq Khan). umar.sahibzada@gmail.com (Muhammad Umar Khayam Sahibzada), tariq.khan@cust.edu.pk (Muhammad Tariq Khan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Abstract
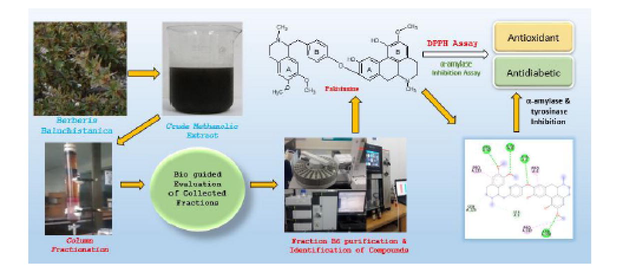
Bioassay based fractionation of methanolic extract of Berberis baluchistanica (Berberidaceae), used traditionally for internal injuries, led to the isolation of known compounds (1–4). The structure of these compounds was elucidated by different spectroscopic analysis and available literature data. Antidiabetic and antioxidant potentials of B. baluchistanica fractions and isolated compounds were evaluated using in vitro alpha- amylase and DPPH assays. The isolated compounds were identified as obamegine (1), pakistanine (2), 8-oxyberberine (3) and baluchistine (4). Obamegine was reported from many other species of this genus but it is first time isolated from B. baluchistanica in present study. Moreover, in vitro pakistanine (2) was found as bioactive lead molecule for hypoglycemic (IC50:40.26 µg/ml) and antioxidant (IC50:14.15 µg/ml) activities compared to acarbose (IC50:33.68 µg/ml) and ascorbic acid (IC50:0.41 µg/ml). To the best of our knowledge, no previous data were available for these biological activities. Additionally, in silico antidiabetic and antioxidant activity of pakistanine against two proteins, α-amylase (-9.7 kcal/mol) and tyrosinase (-8.7 kcal/mol) are reported here for the first time. The molecular docking binding interactions authenticate and support the above-mentioned activities and are helpful in predicting the mechanism of action of pakistanine (2).
Keywords
Alpha amylase assay
Berberis baluchistanica
DPPH assay
Molecular docking
Pakistanine
Tyrosinase
1 Introduction
Baluchistan (Pakistan) is a rich source of medicinal plants due to its diverse landscape and extreme hot and cold climate. Berberis baluchistanica (Berberidaceae), a wild plant found in different areas, is used as fodder and folk medicine to treat internal injuries. Antioxidant potential of crude methanolic extract (CME) and aqueous and organic solvent fractions (chloroform, n-hexane and acetone) of B. baluchistanica roots was reported in literature. Glucose lowering and toxicological effect of crude methanolic extract of B. baluchistanica were also reported (Baloch et al., 2013).
Phytochemical analysis of B. baluchistanica revealed the presence of alkaloids, flavonoids, phenols, saponins and diterpenes in CME and its fractions (Baloch et al., 2013). Literature revealed the presence of various alkaloids (Isoquinoline) pakistanine, pakistanamine (Shamma et al., 1973), quettamine, secoquettamine, dihydrosecoquettamine, baluchistine (Miana et al., 1979), berberine, palmatine and 8-oxoberberine. Polyphenolic compound gallic acid, oleanolic acid a pentacyclic triterpenoid and berberisinol (Pervez et al., 2019) were also isolated from B. baluchistanica. Although various compounds had been isolated from this species and crude extract of roots and stem had been reported to exert significant antioxidant and hypoglycemic activities (Muddassir et al., 2022), but to the best of our knowledge, none of the compounds were evaluated as bioactive agent for antioxidant and hypoglycemic effects. Therefore, the aim of the present study was to search bioactive lead molecule(s) with antioxidant and antidiabetic profile through bioactivity guided fractionation process of B. baluchistanica stem extract and to authenticate its outcomes through in silico molecular docking of isolated compound(s) against two protein targets i.e. alpha-amylase and tyrosinase.
2 Materials and methods
2.1 Plant source
B. baluchistanica stems were identified and collected from Quetta-Baluchistan (Pakistan) with support of Mr. Irshadullah Gondal, District forest officer. The stems were authenticated by Professor Dr. Ghulam Abbas Miana Riphah International University Islamabad, Pakistan. The plant was washed and dried in shade. It was cut into small pieces, ground to fine powder and stored in air-tight container at room temperature.
2.2 Chemicals and drugs
DPPH, resveratrol, acarbose and α-amylase were procured from Sigma Aldrich. Starch, Carboxymethyl cellulose, ascorbic acid (99.8 % Purity) and metformin hydrochloride (99.6 % Purity) were provided by Amson vaccines and pharma, Islamabad. Monobasic potassium phosphate, methanol, dimethylsulfoxide, hydrochloric acid, iodine and potassium iodide were of analytical grade.
2.3 Extraction and bioactivity guided fractionation
B. baluchistanica CME obtained by percolation (soaked in methanol for two weeks) yielded a concentrated dark greenish brown semisolid after vacuum evaporation (Muddassir et al., 2022). For the exploration of most potent biological activity, CME was evaluated for different activities (cytotoxic, antidiabetic, antimicrobial and antioxidant). Significant radical scavenging and hypoglycemic effects amongst tested biological activities set forth a line for further explorations in this regard. 8.0 g of CME was mixed with slurry of silica gel (HF 60, merck 70–230 mesh, 10 × 50 cm) and methanol, dried and loaded on top of glass column as layer, partitioned and eluted with graded mixture(s) of chloroform: methanol. Seven fractions were collected and air dried. Fractions weighing B1 (0.72 g), B2 (0.42 g), B3 (0.78 g), B4 (0.54 g), B5 (1.03 g), B6 (1.36 g) and B7 (1.12 g) were obtained. With the intention of searching most active antidiabetic and antioxidant fraction, all the fractions were screened through α-amylase and DPPH assay respectively. In comparison to other fractions, B5 and B6 exhibited superior α-amylase enzyme inhibition property (Fig. 1). Fraction B6 showed highest antioxidant response IC50:24.90 µg/ml (Table 1). Fraction B6 was preferred for further investigations to isolate potential compounds for hypoglycemic and radical scavenging effects.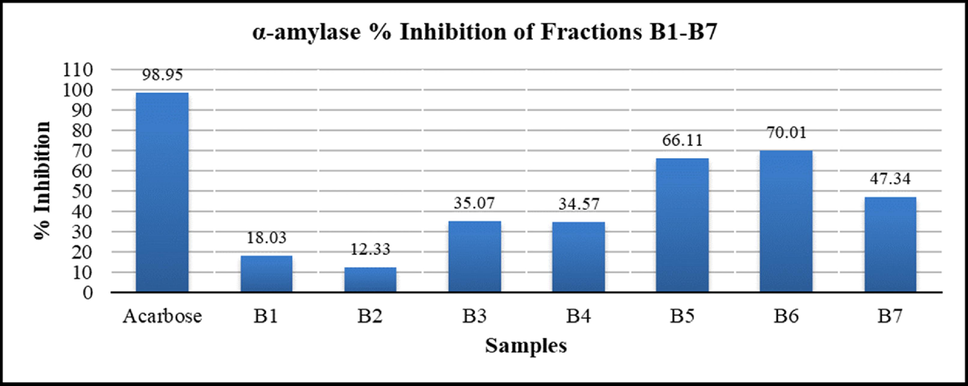
α-amylase Inhibition (%) of B. baluchistanica CME, Fractions (B1-B7) and standard. values calculated as mean ± Standard deviation (n = 3), (P < 0.05).
Sample
Concentration (µg/ml)
IC50 (µg/ml)
25
50
100
250
500
% inhibition
Ascorbic acid
96.34
96.5
96.66
96.82
97.29
0.41
B1
12.1
15.92
33.28
46.82
55.41
102.78
B2
12.42
17.2
32.64
44.59
54.62
116.12
B3
12.9
18.79
34.39
58.6
59.08
88.57
B4
12.26
25.8
44.75
73.41
75.32
83.11
B5
8.76
21.5
35.35
66.56
83.76
168.36
B6
48.09
66.24
79.94
85.35
95.54
24.90
B7
9.08
28.03
51.91
85.67
86.78
82.00
2.4 Isolation of compounds 1–4
Low pressure gradient HPLC with PDA detector (Shimadzu) was utilized to ascertain possible component(s) in B6 fraction. Variable ratios of acetonitrile and water (mobile phase) were applied on reversed phase column (0.46 cm × 25 cm, 5 µm, L1). Samples were scanned between 190 and 800 nm at 1.0 ml/min flow rate. Four peaks/components were eluted at variable retention time (Fig. 2). Subsequently, fraction B6 was subjected to preparative recycling HPLC (LaboACE-LC5060) using silica based reversed phase octadecylsilyl (JAIGEL-ODS-AP) column with particle size 15 µm, pore size 120 Å and surface area 300 m2/g. Components were eluted with mobile phase acetonitrile: water (60:40) at a flow rate of 9.0 ml/min and detected at UV-254 nm wavelength. Upon three repeated 10 ml injections (20 mg/ml of B6 fraction), sub fractions (1–4) were collected as per retention time (Fig. 3). These separated fractions 1–4 were individually dried in vacuum desiccator and corresponded to compounds (1–4). These isolated compounds (1–4) were identified and verified through spectral characterization as obamegine, pakistanine, 8-oxyberberine and baluchistine respectively. Compounds (1–4) were evaluated through in vitro bioactivity based assay to search principal molecule with a potent hypoglycemic and radical scavenging behavior. Pakistanine (2) was found as bioactive lead molecule for hypoglycemic and antioxidant activities.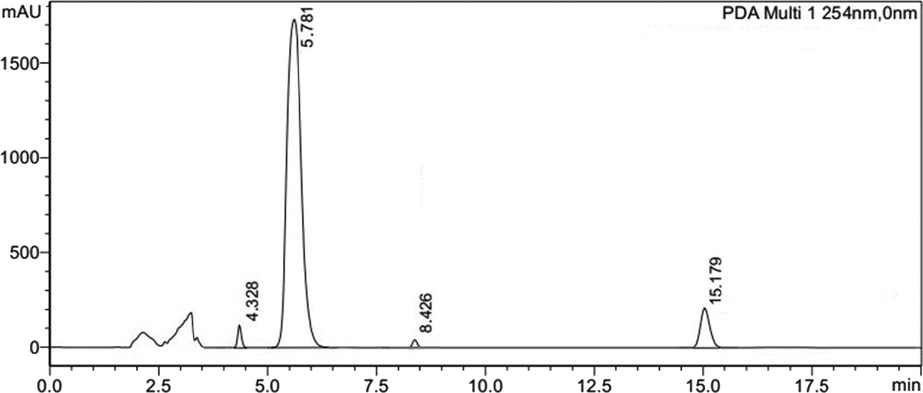
Reversed phase HPLC chromatogram of B6 fraction that was consisting of four peaks with retention time 4.32, 5.17, 8.42 and 15.78 min respectively.
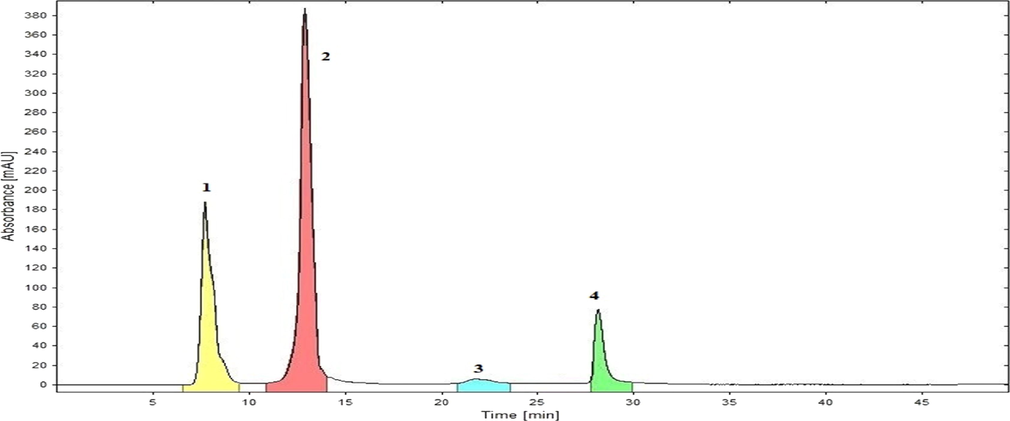
Recycling HPLC chromatogram of B6 Fraction was separated into (1–4) Peaks / sub fractions. These peaks (1–4) were corresponded to compounds 1–4 with retention time 7.6, 12.0, 22.2 and 28.8 min respectively.
2.5 Antidiabetic assay (α − amylase inhibition)
α-amylase inhibition assay was carried out to determine the antidiabetic potential of fractions (1–7) and isolated compounds (1–4) using a standard protocol with slight modifications (Harris, 1992). Phosphate buffer pH 6.7 (10 µl), 10 µl of sample (Fractions, compounds and standard drug) in DMSO, α-amylase enzyme (15 µl) and 0.1 % starch (30 µl) in phosphate buffer pH 6.7 were mixed to prepare a reaction mixture. Separate solution (1000 µg/ml) of fractions (B1-B7) and 25, 50, 100, 200 and 250 µg/ml of compounds (1–4) and standard drug acarbose were used. 96 well microtiter plate was incubated at 50 °C for 30 min to progress enzymatic reaction. 1 M HCl (15 µl) was used to terminate the enzymatic reaction. 0.005 M iodine reagent (80 µl) was added to reaction mixture to react with remaining starch imparting blue color to the solution. Blank solution was prepared by mixing DMSO and phosphate buffer pH 6.7 in equal ratios. Acarbose (0.1 %) was used as positive control, DMSO was utilized instead of sample solution for negative control. Absorbance was measured at 620 nm using microplate absorbance reader in triplicate and results were deduced by following equation expressed as Mean ± SD. % α-amylase inhibition = (S—N)/(B-N) × 100.
Where,
S = Absorbance of sample.
N = Absorbance of negative control.
B = Absorbance of blank.
2.6 Antioxidant assay (DPPH radical scavenging)
DPPH radical scavenging activity was assayed to determine the antioxidant potential of fractions and compounds (1–4) using standard procedure (Obied et al., 2005). DPPH solution (0.032 mg/ml) was prepared in mixture of methanol and water (82:18). Solution of ascorbic acid (reference), fractions and compounds 1–4 (samples) were prepared in methanol (concentration range 25–500 µg/ml). Reference and sample solution (0.2 ml) for each concentration were mixed with 2.8 ml of DPPH solution (in triplicate) in separate glass vials and kept in dark for almost one hour at temperature between 15 and 25 °C. Absorbance of reference, sample and negative control (DPPH solution) was measured at 517 nm using methanol solution as blank. Results (triplicate) were computed by below mentioned equation expressed as Mean ± SD. IC50 values were determined by online calculator (AAT bioquest USA). Results are tabulated in Table 1.
Where,
ADPPH = Absorbance of the DPPH solution.
ATEST = Absorbance of the test solution.
2.7 Characterization of the isolated compounds 1–4
Structure elucidation of compounds (1–4) was carried out using spectroscopic techniques. FTIR spectra were recorded on Bruker Alpha-P, Shimadzu GC–MS quadrupole (Nexis GC-2030) ion trapping and time of flight (TOF) mass spectrometer for obtaining mass spectral data was used. 1H and 13C NMR spectra were measured on Bruker ARX-300 spectrophotometer using TMS as internal standard. DMSO‑d6 was used as solvent at room temperature. Chemical structures of compounds 1, 3 and 4 along with their characterization data and spectra are provided in supplementary material.
2.8 Molecular docking
The molecular docking studies were carried out to analyze the interaction pattern of ligand (pakistanine) with the target proteins alpha-amylase (PDB: 4 W93) and tyrosinase (PDB: 3NM8). The protein data bank was used to access the X-ray crystallographic structure of proteins. The proteins were initially prepared before proceeding for docking by removing the water molecules and ligand from the protein structure. This was done using Discovery Studio 4.0 ( https://www.accelrys.com). Chembiodraw ultra 14.0 was used to draw the ligand and was saved in.mol format. The energy minimization of ligand was done using Chembio3D ultra using mm2 force field. The ligand was saved in pdb format. The pdb files were converted into.pdbqt format using AutoDock Tool version 1.5.6 (Morris et al., 2009) and cubic grid box of ligand and protein interactions was adjusted having dimensions of 45 Å (x, y, z) and 60 Å (x, y, z) for 4 W93 and 3NM8 was generated respectively. PyRx software (Dallakyan and Olson, 2015) was used to determine the binding energies in the performance of docking analysis and the ligand was ranked according to the binding scores. The best poses were saved and visualized using Discovery studio 4.0.
2.8.1 Interaction analysis
The Discovery studio 4.0 ( https://www.accelrys.com) was used to analyze the binding pattern of ligand with the target proteins. The amino acids involved in bonding were visualized and studied.
2.9 Statistical analysis
Αlpha-amylase inhibitory assay was performed in triplicate. Values are stated as mean ± standard deviation (Mean ± SD). Statistical analysis was carried out using ANOVA to relate the mean values of fractions and isolated compounds. Values of p < 0.05 were considered as significant.
3 Results
3.1 Biological activities of fractions (B1-B7) and compounds (1–4)
Fractions (B1-B7) and Compounds (1–4) were evaluated through in vitro bioactivity based assay to search principal molecule with a potent hypoglycemic and radical scavenging behavior. Fraction B6 was found with highest hypoglycemic (% inhibition = 70.01 %) and radical scavenging (IC50 = 24.90 µg/ml) activity as compared to acarbose (% inhibition = 98.95 %) and ascorbic acid (IC50 = 0.41 µg/ml) respectively (Table 1; Fig. 1). Compound 2 (Pakistanine) exhibited significant α − amylase inhibition (IC50 = 40.26 µg/ml) and antioxidant effect (IC50 = 14.15 µg/ml) compared to standard drug acarbose (IC50 = 33.68 µg/ml) and ascorbic acid respectively. IC50 values of compounds 1–4 are tabulated in Table 2..
alpha-amylase % Inhibition
Sample
Concentration / % inhibition
IC50 (µg/ml)
25
50
100
200
250
Acarbose
72.96 ± 0.23
80.08 ± 0.41
88.98 ± 0.28
94.23 ± 0.32
98.60 ± 0.24
33.68
Compound 1
4.21 ± 0.24
10.40 ± 0.28
21.48 ± 0.29
34.63 ± 0.24
44.15 ± 0.24
481.68
Compound 2
31.10 ± 0.14
60.58 ± 0.23
70.72 ± 0.23
85.08 ± 0.34
92.36 ± 0.59
40.26
Compound 3
7.28 ± 0.19
13.47 ± 0.29
21.48 ± 0.29
37.65 ± 0.14
46.91 ± 0.28
1425.14
Compound 4
5.82 ± 0.19
21.27 ± 0.29
30.63 ± 0.23
46.96 ± 0.09
56.94 ± 0.50
219.07
DPPH % Inhibition
Sample
Concentration / % inhibition
IC50 (µg/ml)
25
50
100
250
500
Compound 1
6.65
13.45
22.69
41.29
62.56
833.56
Compound 2
64.12
73.49
87.64
94.61
96.42
14.15
Compound 3
7.94
16.64
25.31
46.84
74.68
2737.08
Compound 4
13.14
23.42
40.25
66.61
82.34
153.53
3.2 Isolation of compounds (1–4)
B6 fraction was analyzed on Low pressure gradient HPLC to ascertain possible component(s). Mixture of acetonitrile and water (60:40) on reversed phase column (0.46 cm × 25 cm, 5 µm, packing L1) was utilized and components were eluted at 1.0 ml/min flow rate, detected at 254 nm. Four peaks/components were obtained at variable retention time (Fig. 2).
Using preparative recycling HPLC, components of fraction B6 were eluted, its sub fractions (1–4) were collected and dried, that were corresponded to compounds (1–4) about 15 mg, 86 mg, 12 mg and 27 mg respectively (Fig. 3).
3.3 Characterization of compound 2
Although obamegine, pakistanine, 8-oxyberberine and baluchistine were identified through different spectral techniques but bioassay based isolated pakistanine (2) shown significant in vitro antidiabetic and antioxidant potential. Therefore spectral analysis of Pakistanine is being reported here (Fig. 4).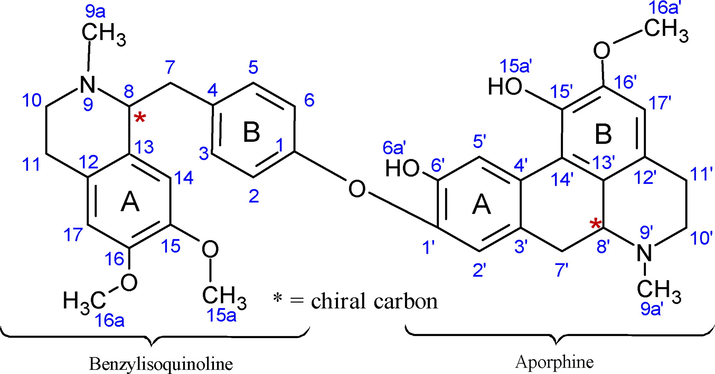
Chemical structure of compound 2 (pakistanine).
White color crystals; Melting point: 153–154 °C; Molecular Formula: C37H40N2O6; Mol weight: 608.7 g/mol; FTIR (cm−1): OH: 3424; CH2 (stretch): 2932; O-CH3: 2845; N-CH3: 2804; C⚌C-C: 1580, 1446; C⚌C aromatic: 1503; Ar-OH (bend): 1360 (Fig. 5). FAB-MS m/z: 608.5 [M + H] +; ESI-TOF m/z: 207 [M − b] +, 402 [M − a] +, 312 [M-(a + b)] –, 296 [b-d] +, 107 [b-e] – (Fig. 6).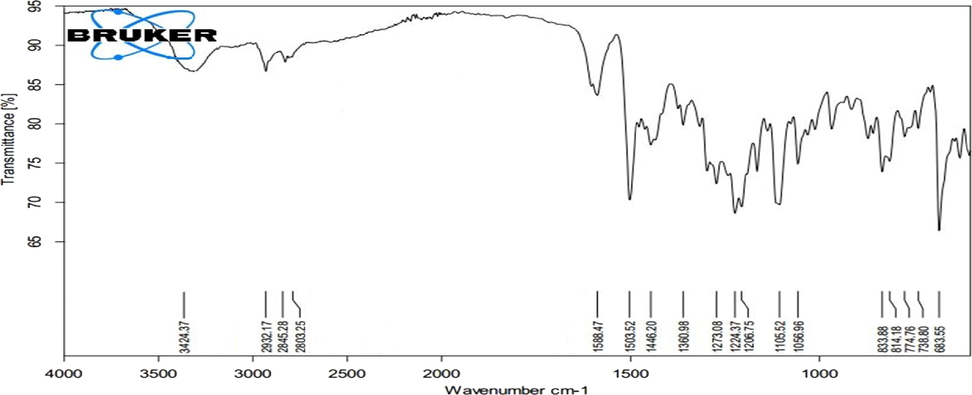
FT-IR spectrum of Pakistanine (2).
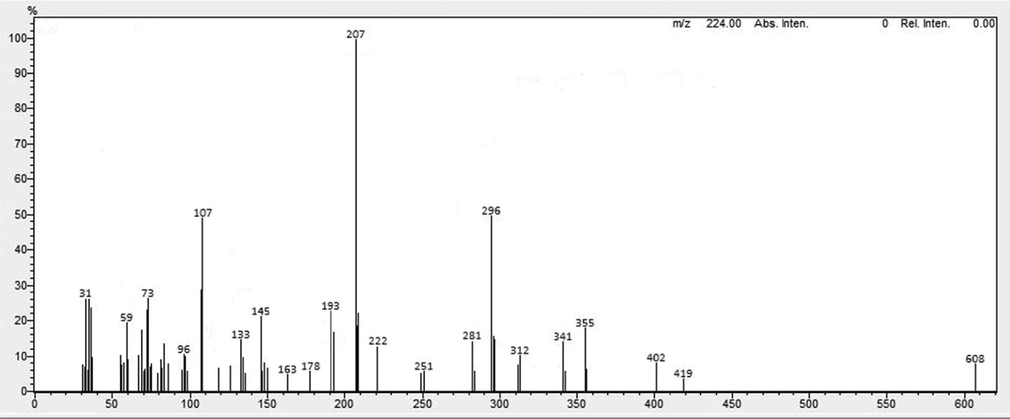
Mass spectrum of Pakistanine (2).
1H NMR (DMSO‑d6): δ8.13 (s, 1H, H5′), δ7.08–7.15 (dd, J = 7.5, 1H, H2-H6), δ6.97 (s, 1H, H2′), δ6.78 (s, 1H, H14), δ6.65 (s, 1H, H17′), δ6.58 (s, 1H, H17), δ5.35 (s, 1H, H6a′, 15a′), δ4.34 (m, 1H, H7′), δ3.81 (s, 3H, H15a), δ3.80 (s, 3H, H16a′), δ3.79 (s, 3H, H16a), δ3.45 (m, 1H, H8), δ2.97–3.00 (m, 2H, H11′), δ2.49–2.93 (m, 2H, H7, H7′ H10′, H11′), δ2.70–2.79 (m, 2H, H10, H11), δ2.43 (s, 3H, 9a′), δ2.35 (s, 3H, 9a).
13C NMR (DMSO‑d6): δ 156.6 (C-1), 149.0 (C-16′), 147.6 (C-15), 147.3 (C-16), 146.9 (C-6′), 145.6 (C-1′), 143.4 (C-15′), 131.9 (C-4), 130.0 (C-3, C-5), 129.1 (C-3), 128.2 (C-13,13′), 127.4 (C-12,12′), 126.8 (C-4′), 121.0 (C-14′), 120.9 (C-2,6), 114.1 (C-5′), 112.3 (C-2′), 111.5 (C-14), 110.7 (C-117′), 64.4 (C-8), 62.5 (C-8′), 56.3 (C-16a′), 56.1 (C-16a), 56.0 (C-15a), 49.3 (C-10′), 46.0 (C-10), 42.8 (C-9a), 42.6 (C-9a′), 39.0 (C-7), 34.5 (C-7′), 26.4 (C-11′), 25.2 (C-11).
In the 1H NMR spectra of pakistanine, benzylisoquinoline moiety showed aromatic protons signals of ring A at δ 7.08–7.15 (dd, J = 7.5), ring B aromatic proton at δ 6.78 and δ 6.58 for position C-14 and C-17 respectively. Aprophine moiety aromatic protons (C-2′, C-17′) as singlet at δ 6.97 and δ 6.65 respectively, whereas unsubstituted benzylic proton of position C-5′ appeared low field at δ 8.13. Six N-methyl protons (9a, 9a′) up fielded as singlet at δ 2.35 and δ 2.43 whereas three methoxy protons shifted downfield at δ 3.79–3.81. Two Hydroxyl protons (6a′, 15a′) as singlet at δ 5.35. Deshielding effect on methine proton (8, 8′) at δ 3.45 and δ 4.34 respectively attributed to the electron withdrawing effect of adjacent nitrogen and further downfield shift of 8′ proton (δ 4.34) resulted from cyclic ring. Methylene protons showed signals at δ 2.49–3.00 (m, J = 7.1 & 7.2). 13C NMR (DMSO‑d6) spectrum displayed signals of two N-methyl δ 42.6, 42.8 and three methoxy carbon at δ 56.0–56.3. Phenolic carbon (C-6′) appeared upfield at δ 146.9 due to shielding effect of aryl ether substituent and due to impact of alkyl ether substituent shielding shifted C-15′ values further upfield at δ 143.4. Ether linkage between C-1 and C-1′ signaled low field δ 156.6 and δ 145.6 due to para substituent benzylic and ortho substituted phenolic groups respectively (Pavia et al., 2014). Findings of spectral data (IR, Mass, H NMR) were consistent with the literature reported data. However for authentication and more detailed spectroscopic prospective, 13C NMR (pakistanine) spectra was recorded and data is reported in this research for the first time. (1H NMR and 13C NMR spectra of compound 2 provided in supplementary materials as fig. S8 and S9 respectively).
3.4 Molecular docking
The protein–ligand docking analysis of pakistanine (2) against target proteins was performed to evaluate the best binding affinity of ligand–protein complex based on docking scores.
3.4.1 Docking studies on α-amylase (code 4w93)
Human pancreatic alpha-amylase (code 4w93), a hydrolytic enzyme serves as a vital and predominant source for maintaining blood glucose levels through dietary carbohydrate hydrolysis (Harris, 1992).
Binding orientation of ligand with protein α-amylase is shown in Fig. 7, in ligand-alpha amylase complex, the ligand manifested lowest binding score of − 9.7 kcal/mol. The interaction analysis revealed that in protein amylase, the amino acid residues ARG92, GLN8, THR6 and ARG398 established four key stable hydrogen bonds in protein pocket with methoxyl group and therefore a stable ligand–protein complex as well as an efficient amylase inhibition is concluded. Residues PRO4, PRO223 and PRO332 were observed in the alkyl and pi-alkyl complexation with methoxy and benzene functionalities of benzylisoquinoline and aporphine of compound (2). RMSD (Root-mean-square deviation) for ligand-alpha amylase complex was found as 1.6. Lowest energy binding score is shown in Table 3.. RMSD upper bound describes the matching of atoms in one conformation with itself and in the other probable conformation, disregarding any symmetry. RMSD lower bound compares one atom in a conformation with the nearest atom of the similar type in other conformation.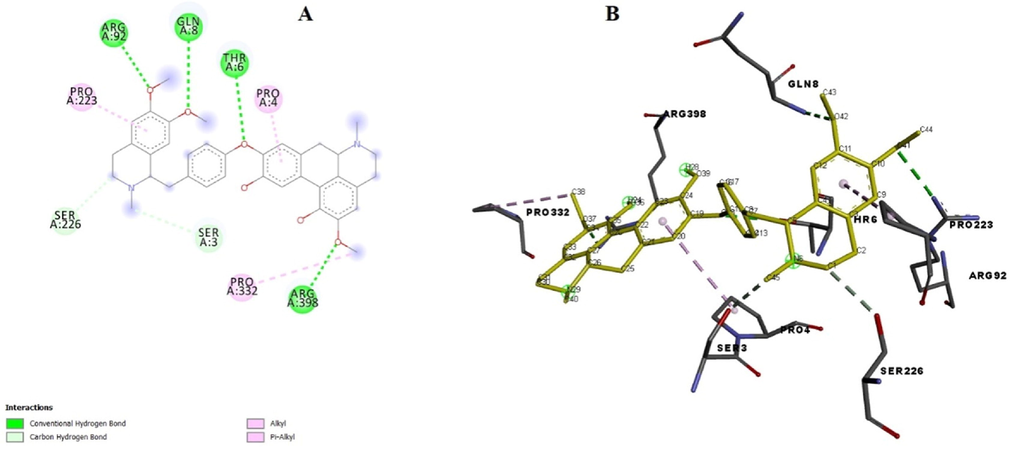
(A) Binding poses of ligand with the target alpha-amylase showing surface interaction. (B) Amino acid interaction of ligand with the protein.
Ligand
Binding Affinity (kcal/mol)
RMSD upper bound
RMSD lower bound
4w93_aamylase_MA
9.7
1.607
0.507
4w93_aamylase_MA
9.1
2.707
1.622
4w93_aamylase_MA
9.0
40.77
39.317
4w93_aamylase_MA
9.0
11.887
1.867
4w93_aamylase_MA
8.8
40.686
37.686
4w93_aamylase_MA
8.6
41.613
38.565
4w93_aamylase_MA
8.6
11.695
2.638
4w93_aamylase_MA
8.6
36.833
31.86
4w93_aamylase_MA
8.5
17.802
7.864
3.4.2 Docking studies on protein tyrosinase (code 3NM8)
The enzyme tyrosinase is a metalloenzyme group of polyphenol oxidases that occurs in various organisms and performs some specific functions of oxidation (Videira et al., 2013). Multiple natural and synthetic based potent tyrosinase inhibitors have been reported during the last decade (Lee et al., 2016). In order to assess the interaction between the pakistanine (2) and tyrosinase (code 3NM8) molecular docking study was performed using the PyRx software. The lowest binding score for oxidative protein tyrosinase was − 8.7 kcal/mol (RMSD: 1.87). In protein tyrosinase the amino acid PHE258 showed key conventional hydrogen bonding with hydroxyl group and pi-pi contact with benzene of aporphine moiety, moreover, van der Waals interactions with GLY256 for same group were also observed. Residue ASP264 contributed pi-anion interaction with the benzene ring of aporphine (Fig. 8). Residue LYS254, LYS32, PRO51 established alkyl and pi-alkyl interaction with methoxy and benzene group of benzylisoquinoline and aporphine moieties of the ligand. These interactions established efficient binding of ligand and suggested stable protein–ligand complex. Binding affinity score is shown in Table 4..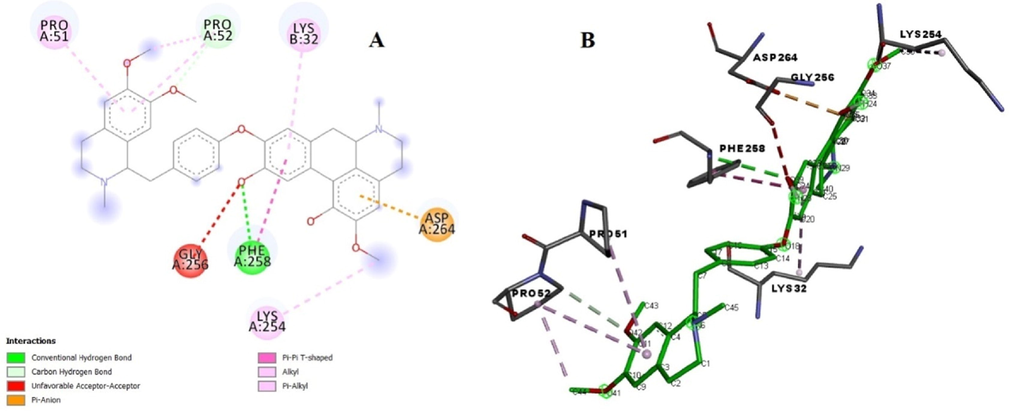
(A) Binding poses of ligand with the target protein tyrosinase showing surface interaction. (B) Amino acid interaction of ligand with the protein.
Ligand
Binding Affinity (kcal/mol)
RMSD upper bound
RMSD lower bound
3nm8_MA
8.9
1.87
1.32
3nm8_MA
8.7
29.452
26.505
3nm8_MA
8.7
41.715
38.786
3nm8_MA
8.6
13.645
6.873
3nm8_MA
8.6
29.622
25.993
3nm8_MA
8.6
8.431
4.946
3nm8_MA
8.3
37.32
31.598
3nm8_MA
8.2
20.28
12.691
3nm8_MA
8.1
14.071
2.824
4 Discussion
Biomolecules are prone to free radicals formed either by cellular metabolism, oxidative stress or extrinsic factors, consequentially causing various ailments like cancer, neurodegenerative, cardiovascular diseases and diabetes mellitus (Bland, 1995). Antioxidant compounds nullifies free radical formation in order to safeguard oxidative deterioration. Carcinogenic and hepatotoxicity of synthetic antioxidants like butylated hydroxy toluene (BHT) and butylated hydroxy anisole (BHA) were reported (Ito et al., 1986), whereas consumption of natural antioxidants reduce the risk of diabetes mellitus as compared to synthetic antioxidants (Rafieian-kopaei et al., 2015). Plants with antidiabetic activity also possess antioxidant potential (Hulbert et al., 2005; Rafieian-Kopaie, and Nasri, 2012) and the dire need to identify natural antioxidants of botanical source is vital for human health. In this research, bioassay guided fractionation of B. baluchistanica stem extract provided four known alkaloids obamegine (1), pakistanine (2), 8-oxyberberine (3) and baluchistine (4). Obamegine is first time outcome of this species, however this compound had been previously reported from other species of genus berberis. Results of the present studies disclosed the antioxidant and antidiabetic aspect of bioactivity based isolated pakistanine (2) from B. baluchistanica. Likely mechanism of pakistanine in reduction of blood glucose level may be due to decrease in oxidative stress. Targeted therapies have been proven to boost up the standard of life in either case of disease, therefore extensive studies which address targeted diabetic regimes have opened research gateways; in this research two target proteins (alpha amylase and tyrosinase) were examined through docking. In order to verify, validate, identify and evaluate the key binding interactions and types of binding pockets of proteins available in the 4w93 and 3NM8, docking of the ligand (pakistanine) with these proteins was executed through AutoDock Tool (version 1.5.6). Based on the docking results, RMSD in both of the cases was calculated as < 2.0, poses are usually regarded “near-native” with RMSD ≤ 2.0 Å. Strong binding affinity of pakistanine (2) with both the targeted proteins suggested it as stable and efficient alpha amylase and tyrosinase natural inhibitor or significant antidiabetic and antioxidant bioactive agent respectively. Additionally, pakistanine exhibited the remarked antidiabetic and antioxidant activity which can be associated to its phenol ring. Phenolic compounds generally demonstrate an antioxidant or pro-oxidant effect according to their concentration as well as due to presences of the phenolic ring by donation of hydrogen atoms or electrons (Ferguson, 2001).
5 Conclusion
Oxidative degeneration of human cells can be shielded with antioxidants which can scavenge free radicals produced during the process of cellular breakdown, oxidative stress and extrinsic factors. Oxidative stress is a key factor in development of diabetes mellitus. Carcinogenic and hepatotoxic adverse effects of synthetic antioxidants trigger the utilization of antioxidants of natural origin. Bioassay based fractionation and separation of B. baluchistanica crude extract brought about the isolation of obamegine (1), pakistanine (2), 8-oxyberberine (3) and baluchistine (4). However the natural product pakistanine (2) was found as the preferential in vitro antioxidant and hypoglycemic agent for the first time. The docking score data would be promising to predict the inhibitory approach in fast and precise ways in future.
6 Author’s contributions
All the authors contributed equally in the preparation of this manuscript. All the authors have revised the manuscript as suggested by the reviewer and approved the final draft for submission.
Acknowledgements
We are thankful to Islamic international university Islamabad, Pakistan for the GC-MS analysis. Riphah international university for preliminary work and Recycling HPLC. Amson Vaccines and Pharma Islamabad for performing biological activities. The authors also like to thank Taif University, Taif, Saudi Arabia, for their support (Taif University Researchers Supporting Project number: TURSP-2020/80).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- In-vitro antileishmanial, cytotoxic, antioxidant activities and phytochemical analysis of Berberis baluchistanica roots extracts and its fractions. J. Phytopharm.. 2013;4:282-287.
- [Google Scholar]
- Oxidants and antioxidants in clinical medicine: past, present and future potential. J. Nutr. Environ. Med.. 1995;5:255-280.
- [CrossRef] [Google Scholar]
- Dallakyan, S., Olson, A.J., 2015. Small-Molecule Library Screening by Docking with PyRx, in: Chemical Biology. Springer, pp. 243–250. https://doi.org/10.1007/978-1-4939-2269-7_19.
- Role of plant polyphenols in genomic stability. Mutat. Res. Mol. Mech. Mutagen.. 2001;475:89-111.
- [CrossRef] [Google Scholar]
- Classification of diabetes mellitus and other categories of glucose intolerance. Int. Textb. diabetes Mellit. 1992:3-18.
- [Google Scholar]
- Dietary fats and membrane function: implications for metabolism and disease. Biol. Rev.. 2005;80:155-169.
- [CrossRef] [Google Scholar]
- Studies on antioxidants: their carcinogenic and modifying effects on chemical carcinogenesis. Food Chem. Toxicol.. 1986;24:1071-1082.
- [CrossRef] [Google Scholar]
- Natural, semisynthetic and synthetic tyrosinase inhibitors. J. Enzyme Inhib. Med. Chem.. 2016;31:1-13.
- [CrossRef] [Google Scholar]
- Baluchistine, a new bisbenzylisoquinoline alkaloid. Experientia. 1979;35:1137-1138.
- [CrossRef] [Google Scholar]
- AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem.. 2009;30:2785-2791.
- [CrossRef] [Google Scholar]
- Comparative studies of anticancer, antimicrobial, antidiabetic, antioxidant activities of Daphne oleoides and Berberis baluchistanica extracts native to Pakistan. Pak. J. Pharm. Sci. 2022:649-656.
- [CrossRef] [Google Scholar]
- Investigation of Australian olive mill waste for recovery of biophenols. J. Agric. Food Chem.. 2005;53:9911-9920.
- [CrossRef] [Google Scholar]
- Introduction to spectroscopy. Cengage Learning; 2014.
- Antimicrobial and antioxidant potential of Berberisinol, a new flavone from Berberis baluchistanica. Chem. Nat. Compd.. 2019;55:247-251.
- [CrossRef] [Google Scholar]
- Antioxidant plants and diabetes mellitus. J. Res. Med. Sci.. 2015;20:491.
- [CrossRef] [Google Scholar]
- Pakistanine and Pakistanamine, two new dimeric isoquinoline alkaloids. J. Am. Chem. Soc.. 1973;95:5742-5747.
- [CrossRef] [Google Scholar]
- Videira, I.F. dos S., Moura, D.F.L., Magina, S., 2013. Mechanisms regulating melanogenesis*. An. Bras. Dermatol. 88, 76–83. https://doi.org/10.1590/S0365-05962013000100009.
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.104221.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1







