Plant antiviral compounds containing pyrazolo [3,4-d] pyrimidine based on the systemin receptor model
⁎Corresponding author at: State Key Laboratory of Green Pesticide, Key Laboratory of Green Pesticide and Agricultural Bioengineering, Ministry of Education, Guizhou University, Huaxi District, Guiyang 550025, China. jwu6@gzu.edu.cn (Jian Wu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Introduction
The development of small molecule modulators rooted the plant resistance mechanism represents a compelling strategy for improving plant resistance and fortifying food security. Systemins induced by wounds can activate a wide range of plant resistance responses, but compounds that regulate the systemin signaling pathway have yet to be found.
Objectives
This study aimed to provide a strategy for the development of small molecule modulators based on virtual screening of systemin receptors.
Methods
A systemin receptor model was established for virtual screening, leading to the identification of chemical structure with high binding affinity through molecular docking. Subsequently, the synthesized compound underwent evaluation for its potential to enhance plant resistance against viruses through frictional inoculation. The underlying mechanism of the target compound was analyzed using proteomics.
Results
Through virtual screening, a promising active compound, F1, was identified and further optimized. Molecular docking showed that they had the ability to bind SR160. Some of the targeted compounds exhibited potential in enhancing plant resistance against Tobacco mosaic virus (TMV) and Cucumber mosaic virus (CMV). Notably, compound F26 showed satisfactory activity against TMV with an EC50 value of 70.6 μg/mL. The analysis of plant response following treatment with F26 indicated that it may reinforce the plant’s defense mechanisms against viruses by augmenting the wound signaling pathway.
Keywords
Defense response
Novel agrochemical
Pyrazolo[3,4-d] pyrimidine
Hydrazine
Virtual screening
Systemin receptor
1 Introduction
Plants inhabit complex environments, inevitably contending with pests, weeds, pathogens and other adverse environment factors. To adapt to these challenges, plants have established defense mechanisms to cope with biotic stresses (Zhang et al., 2022). These mechanisms include the immune response with salicylic acid (SA) as the key signal and the defense response with jasmonic acid (JA) as the key signal. Which signaling pathway plants take depends on the perceived external stress conditions. For example, the SA signal can be induced by pathogen invasion (Han, 2019), while the JA signal can be induced by insect attack (Song et al., 2017). Phytophagous insects feed on the leaves of plants, which is a kind of damage to the plants. Systemins are synthesized in damaged leaves and are transmitted downward by plants as a wound signal to activate defense responses, including the synthesis of JA and the production of protease inhibitors (PIs) (Schilmiller and Howe, 2005). Although plants have distinct response mechanisms to different biological stresses, they share many physiological similarities. This includes the production of oxidative stress, the synthesis of secondary metabolites such as lignin, toxic phenols, and flavonoids, as well as the production of plant protectors (Appu et al., 2021; War et al., 2012). Consequently, the defense responses induced by the immune signal like SA or the defense signals like JA have broad-ranging effects on biotic stresses (Santino et al., 2013).
Diseases caused by plant viruses pose a serious threat to crop yields. Developing drugs that directly target plant viruses is a challenge. Ningnanmycin (NNM), which has been widely used in China, was only about 50–60 % effective against plant viruses in the field (Y. J. Wang et al., 2018; Zhang et al., 2023a). It is a feasible strategy for agricultural chemists to mitigate the damage caused by plant viruses by improving the defense capability of plants. Some compounds with plant activity have been derived based on the structure of JA and SA (Frackenpohl et al., 2023). Furthermore, certain commercial agents like Benzothiadiazole, Dufulin, Vanisulfane, have been designed to enhance plant immune responses against pathogen invasion through the SA immune pathway in plants have been developed (Huang et al., 2023; Zheng et al., 2023). Coronatine (COR), produced by the plant pathogen Pseudomonas Syriana, has been used as a substitute for JA in agricultural production (Feys et al., 1994; Yan et al., 2009). However, bioactive molecules acting on the basis of systemin signaling pathway have not been identified.
Systemins were first identified in tomato and involved in the regulation of tomato defense response to insect feeding (Scheer and Ryan, 2002). They are referred to as signal peptides with a sequence of 18 amino acids that induce plant defense responses, but the amino acid sequences are not consistent among different plants (Bowles, 1998; H. Zhang et al., 2020). The systemin receptors belong to the Leu-rich repeat receptor-like kinase (LRR-RLK) family (Chakraborty et al., 2019; L. Wang et al., 2018). Upon activation by systemin, these receptors trigger a series of signaling events that include the accumulation of reactive oxygen species, activation of phosphatase, JA synthesis and production of PIs (Sun et al., 2011; H. Zhang et al., 2020). PIs, which are regulated by wound signaling, have a toxic effect on pests (Hellinger and Gruber, 2019). This is also one of the typical features of wound signaling responses (Pearce, 2011). Interestingly, the effect of systemin activation extend beyond defense responses against pest. Systemin treatments have been shown to enhance plant tolerance to salt stress and increase resistance to microorganisms (Cirillo et al., 2022; Molisso et al., 2020; Pastor-Fernández et al., 2022; Zhang et al., 2018). Therefore, it is possible to enhance plant defense against plant viruses by activating the systemin signaling pathway.
As traditional drug research becomes slower and more expensive, seeking the help of cutting-edge technology becomes crucial. Computer aided drug design (CADD) has been widely used in drug development. Compared to traditional methods like random screening and high-throughput screening, virtual screening stands out for its good targeting, negligible side effects, and high efficiency. It is advantageous for shortening drug development cycles and budgets, making it an attractive choice for developing promising new compounds (Q. Li et al., 2023; Z. Li et al., 2023). In this study, we utilized SR160, the first identified systemin receptor from tomato, as the basis for constructing the target model. By leveraging chemical structure libraries for virtual screening, we aimed to identify chemical structures with high binding potential against the systemin receptor. The invasion of plant viruses induces immune signaling pathways in plants without affecting wound signaling pathways (K. Zhang et al., 2020). The defense response triggered by systemin is closely linked to the wound signaling system, making it effective not only against phytophagous insects but also pathogenic microorganisms. Therefore, the differential changes in wound signaling between treatments can be used to determine whether the target compounds have successfully activated the wound signaling pathways.
2 Experimental
2.1 Systemin receptor model and virtual screening
The structural characteristics of plant systemin receptors have yet to be determined. In this study, the 3D structures of SR160 and SYR1 were derived from AlphaFold predictions (AF-Q8L899-F1-model_v4 and AF-A0A3Q7FKC1-F1-model_v4). Additionally, the crystal structure of BRI1 (PDB ID:4M7E) was sourced from the RCSB Protein Database Sun et al., 2013). Virtual screening was conducted in the Molecular Operating Environment (MOE 2019) software, which involved several steps such as protonation, addition of missing atoms, complementation of missing group, energy minimization, energy optimization, and the application of the Amber 14: EHT force field. The virtual screening process was performed twice. Initially, the DO1100 database (https://www.tsbiochem.com/), comprising 43,417 compounds, was used for molecular docking analysis. Subsequently, a pharmacophore model was developed based on the outcomes of the initial screening, and molecular docking simulations were conducted for a series of designed compounds. The results were then analyzed using MOE and PyMOL software tools.
2.2 Synthesis of target compounds
2.2.1 Chemicals
The required reagents for the experiment were purchased from TCI (Tokyo, Japan). Solvents were purchased from Accela (Shanghai, China). Solvents and reagents were used without further purification and drying.
2.2.2 Instruments
Melting points were determined via X-4 binocular microscope melting point apparatus (Beijing Tektronix Instruments Co., Ltd., China; unadjusted). The 1H, 13C, and 19F nuclear magnetic resonance (NMR) spectra of the target compounds were recorded on a JEOL ECX 500 NMR (JEOL Ltd., Tokyo, Japan) or an AVANCE III HD 400M NMR (Bruker Corporation, Fallanden, Switzerland) in CDCl3 or DMSO‑d6 solutions. High-resolution mass spectra (HRMS) were obtained with a Thermo Scientific Q Exactive (Thermo Scientific, Missouri, USA) mass spectrometer.
The synthesis route of the target compounds is shown in Fig. 3, and the detailed synthesis method can be found in the Supplementary Method.
2.3 Response of plants
2.3.1 Plant materials
Nicotiana glutinosa seeds are uniformly spread on a substrate soil and placed in an artificial climate chamber maintained at a light intensity of 10,000 Lux, temperature of 28 °C, and humidity level of 80 % for cultivation. Daily watering was carried out, and the tobacco seedlings were transplanted once they reached a height of approximately 2–3 cm. The cultivation process continued for approximately 40 days until the tobacco plants reached the 5–6 leaf stage, indicating readiness for further use. Chenopodium amaranticolor was also grown following a similar cultivation protocol as Nicotiana glutinosa.
The planting procedure for Nicotiana tabacum K326 closely resembles that of Nicotiana glutinosa. For experimental treatment and sampling, Nicotiana tabacum K326 strains at the 6-leaf stage were selected and evenly divided into four treatment groups: CK (healthy plant), CK + TMV (negative control), NNM + TMV (positive control), and F26 + TMV. Each treatment group consisted of three replicates. A formulated material (4 mL) was evenly applied to the entire leaf surface (Preparation of compound solution: accurately weighing 2 mg of compound in a 15 mL centrifuge tube, adding 40 μL of dimethyl sulfoxide to dissolve it, and adding 4 mL of 1 % Tween 80 to form a solution of 500 μg/mL.). following inoculation of the plants 24 h later, all treatment groups were cultured in an artificial incubator set at 10,000 Lux of light and a temperature of 28 ℃. Samples were collected on the 1st, 3rd, 5th, and 7th days after the plants were inoculated with the TMV. These samples were rapidly frozen using liquid nitrogen and then stored at −80 °C for future analysis.
2.3.2 Antiviral bio-assay against TMV and CMV
The TMV was extracted and purified following the method by Gooding (Song et al., 2005; Wu et al., 2017). The half- leaf spot method was utilized to assess compound‘s ability to induce plant antiviral activity. Plants were treated with the compounds and inoculated with the plant virus 24 h later, with further details provided in the Supplementary Method. The evaluation method for the CMV mirrored that of the TMV as described earlier. The CMV was used instead of the TMV, and the host plant employed was Chenopodium amaranticolor.
2.3.3 Pot experiments in vivo
Prepare a 500 μg/mL concentrated solution of the drug using a 1 % Tween 80 solution. Subsequently, select Nicotiana glutinosa leaves at the six-leaf stage, and uniformly apply the prepared drug solution to the right side of the leaves as the treatment group, while apply a 1 % Tween 80 solution to the left side as the blank control. After 24 h of compounds treatment, sprinkle silicon carbide on the leaves, gently rub to mechanically infect the plants with TMV. After 30 min, wash off the residual silicon carbide from the leaves, then transfer the plants to an artificial climate chamber maintained at a temperature of 28 °C, humidity of 80 %, and light intensity of 10,000 Lux for continued growth. 3–4 days later, observe and record the virus symptoms on the tobacco leaves, with each experiment being repeated three times (Y.J. Wang et al., 2018; Wu et al., 2016; Zhang et al., 2023b). A pot experiment of CMV infection followed a similar treatment scheme as TMV, with the difference being the replacement of the host plant with Chenopodium amaranticolor.
2.3.4 Determination of defense enzyme activity
The defense-related enzyme activities of superoxide dismutase (SOD) and polyphenol oxidase (PPO) were assessed using the standard assay protocols outlined in the enzyme analysis kit instructions (Suzhou Kemin Bioengineering Research Institute, China).
2.3.5 Differential expression protein analysis
Standard methodologies described in existing literature were employed for protein extraction, peptide digestion, LC-MS/MS data acquisition, protein identification and bioinformatics analysis. More details are available in the Supplementary Method. This work was carried out in cooperation with APTBIO (Shanghai, China).
2.3.6 RNA extraction and RT-qPCR analysis
RNA extraction: The RNA extraction process involved comprehensive sample processing. Following the addition of chloroform and subsequent centrifugation, distinct layers comprising a supernatant, middle, and organic layer were formed. Notably, the RNA was primarily concentrated in the upper aqueous phase. Subsequently, the collection of the supernatant layer allowed for isopropanol precipitation, enabling the recovery of Total RNA.
cDNA synthesis: In an RNase-free centrifuge tube, a blend of 16 μL RNase-free ddH2O, 4 μL 4*gDNA wiper Mix, and 1 μL template RNA was incubated at 42℃ for 2 min. Subsequently, the 5 × HiScriptII qRT SuperMix II was introduced into the mixture, followed by reverse transcription conducted in a PCR machine.
RT-qPCR analysis: β-actin was used as the endogenous control. The experimental procedures for RT-qPCR were conducted in accordance with the manufacturer's instructions, and the resulting data were analyzed using the 2−ΔΔCt method.
2.3.7 Statistical analysis
The statistical analysis of the antiviral activity data presented in Tables 2 and 3, as well as the defense enzyme activity data illustrated in Fig. 5, was conducted using SPSS. The antiviral activity data provided in Tables 2 and 3 represent the means ± standard deviations of three replicates. Significant differences were analyzed using one-way ANOVA and Duncan's test, and significant difference results were marked with lowercase letters, with different lowercase letters indicating significant differences (P < 0.05). The RT-qPCR data were analyzed using GraphPad Prism 8.0.2, with statistical analysis conducted through Student's t-test.
| Scores | Number of compounds |
|---|---|
| <−8.5 | 59 |
| <−8.0 | 426 |
| <−7.5 | 1805 |
| <−7.0 | 4513 |
The score is the result of molecular docking performed by MOE. A lower value of it means that the small molecule ligand is more likely to bind to the target protein.
| Compounds | Anti-CMV protection effect (%) | Anti-TMV protection effect (%) | Compounds | Anti-CMV protection effect (%) | Anti-TMV protection effect (%) |
|---|---|---|---|---|---|
| F1 | 33.0 ± 1.2 o-q | 55.1 ± 4.1 i-l | F24 | 48.6 ± 1.0 g-k | 26.3 ± 1.1 s-t |
| F2 | 62.3 ± 2.4 b-c | 40.5 ± 4.0o-p | F25 | 37.6 ± 4.6 m-o | 23.0 ± 2.9 t |
| F3 | 59.4 ± 4.6 c-d | 54.5 ± 1.0 i-l | F26 | 50.7 ± 4.6 e-i | 79.3 ± 2.1 a |
| F4 | 37.7 ± 3.0 m-o | 67.4 ± 3.4 d-f | F27 | 42.2 ± 4.8 k-n | 63.9 ± 3.2 e-h |
| F5 | 52.1 ± 4.7 d-h | 54.9 ± 4.7 i-l | F28 | 54.9 ± 4.0d-g | 51.1 ± 4.0 j-m |
| F6 | 39.3 ± 4.6 l-o | 60.7 ± 3.7 f-i | F29 | 23.4 ± 4.6 r-u | 57.1 ± 3.4 h-j |
| F7 | 45.3 ± 4.9 h-l | 54.4 ± 4.7 i-l | F30 | 62.7 ± 5.0 b-c | 54.3 ± 4.4 i-l |
| F8 | 36.0 ± 0.6 n-p | 49.1 ± 4.3 k-n | F31 | 30.3 ± 4.7 p-r | 43.2 ± 4.7 n-p |
| F9 | 29.3 ± 4.2 p-s | 30.2 ± 4.1 r-s | F32 | 55.1 ± 1.4 d-g | 44.9 ± 0.2 m-p |
| F10 | 55.8 ± 5.0 c-g | 55.7 ± 3.7 i-k | F33 | 52.3 ± 1.4 d-h | 33.7 ± 2.9 q-r |
| F11 | 66.6 ± 5.0 b-c | 59.1 ± 3.8 g-i | F34 | 44.0 ± 4.8 i-m | 31.1 ± 4.5 r-s |
| F12 | 49.4 ± 0.6 f-k | 48.0 ± 2.9 l-n | F35 | 42.9 ± 4.8 h-l | 46.7 ± 4.4 m-o |
| F13 | 42.8 ± 4.3 j-n | 46.6 ± 5.0 m-o | F36 | 56.4 ± 2.3 c-f | 64.2 ± 4.6 e-h |
| F14 | 43.1 ± 5.0 j-n | 38.6 ± 2.9 p-q | F37 | 36.3 ± 3.0 n-p | 77.0 ± 5.0 a-b |
| F15 | 21.7 ± 4.9 t-v | 46.4 ± 3.5 m-o | F38 | 48.5 ± 1.0 g-k | 81.0 ± 4.5 a |
| F16 | 15.5 ± 3.6 v | 46.4 ± 4.9 m-o | F39 | 54.7 ± 3.8 d-g | 62.9 ± 1.3 e-h |
| F17 | 70.9 ± 3.4 a | 44.5 ± 4.8 m-p | F40 | 37.9 ± 4.7 m-o | 71.9 ± 2.3 b-d |
| F18 | 36.3 ± 3.0 n-p | 68.2 ± 4.6 c-e | F41 | 28.0 ± 4.8 q-t | 58.9 ± 3.5 g-i |
| F19 | 54.4 ± 1.9 d-g | 78.4 ± 0.9 a-b | F42 | 22.3 ± 4.3 s-u | 58.8 ± 3.2 g-i |
| F20 | 21.0 ± 5.0 u-v | 49.8 ± 4.5 k-n | F43 | 28.1 ± 4.5 q-t | 65.4 ± 4.7 d-g |
| F21 | 49.9 ± 0.3 f-j | 16.7 ± 5.1 u | F44 | 26.5 ± 4.5 q-u | 63.7 ± 4.1 e-h |
| F22 | 28.8 ± 5.0 q-s | 64.4 ± 3.3 e-g | F45 | 53.3 ± 1.0 d-g | 28.7 ± 5.0 r-t |
| F23 | 30.1 ± 3.3 p-r | 60.0 ± 4.3 g-i | NNM | 57.4 ± 2.0 c-e | 74.4 ± 1.9 a-c |
The data in the table is the average value of three repetitions ± standard error; A one-way ANOVA followed by Duncan's test was used for significant differences at P < 0.05, marked with lowercase letters. Different lowercase letters indicate significant differences.
| Compounds | EC50 of Anti-TMV activity (μg/mL) |
|---|---|
| F19 | 78.8 |
| F26 | 70.6 |
| F37 | 76.5 |
| F38 | 74.6 |
| F40 | 115.5 |
| NNM | 86.5 |
EC50 of some compounds with good antiviral activity. The value of EC50 were calculated using linear regression equations. The concentrations used in linear regression equations were 31.25 μg/mL, 62.5 μg/mL, 125 μg/mL, 250 μg/mL and 500 μg/mL, respectively. NNM is a commercial antiviral agent, Ningnanmycin.
3 Results and discussion
3.1 Target protein model and virtually screening
To date, only two systemin receptors, SR160 and SYR1, have been studied, both of which belong to the leucine-rich repeat receptor kinases. SR160, the first systemin receptor isolated from tomato, is highly homologous to brassinolide (BR) receptor kinase BRI1 (Scheer and Ryan, 2002). BR and systemin can share receptors, but BR does not activate the systemin responses (Malinowski et al., 2009). SYR1 was also identified from tomato as a true systemin receptor (L. Wang et al., 2018; H. Zhang et al., 2020). For virtual screening, reliable protein structure modeling is essential to improve the accuracy of the results (Hassan Baig et al., 2016; Macalino et al., 2015). Therefore, SR160 is more advantageous than SYR1 as a target model for the drug discovery because the predictive model of SR160 can refer to the protein crystal structure of BRI1 (Supplementary Fig. 1). The space of positive ligand sites is another prerequisite for obtaining more accurate drug structures. It seems extremely difficult to chemically mimic the structure of systemins and activate the systemin receptors, because systemin is a peptide. However, Leucine-rich repeat kinase 2 (LRRK2), a member of the leucine-rich repeat kinase family, has been used as a key target protein for the development of drugs to treat Parkinson’s disease (Dorsey et al., 2018). The kinase domain is an important region for drug activity (Zhu et al., 2024). Interestingly, the kinase domains of LRRK2 and SR160 are highly similar, and the drug binding pocket of LRRK2 is preserved in SR160 (Fig. 1). With the idea of pharmaceutical development, the kinase domain of SR160 was used as a potential ligand binding space for virtual screening.
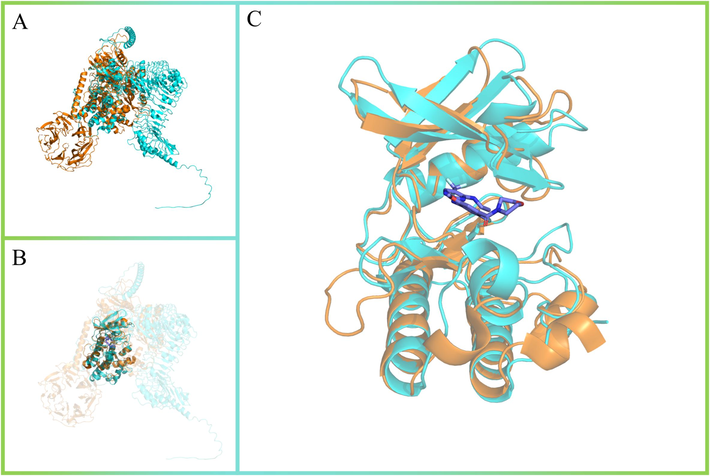
-
Structural comparison of kinase domains between LRRK2 and SR160. (A) The overall 3D structure of LRRK2 (PDB:8U7H) and SR160 (AF-Q8L899-F1-model_v4). The backbone of protein was rendered in tube. LRRK2 was colored in orange and SR160 was colored in cyan. (B) The kinase domains of LRRK2 and SR160 were highlighted. (C) The binding packet of LRRK2-GEN7915 complex.
The results of molecular docking are scored based on energy, with lower scores indicating more stable interactions (Table 1). Among the top ranked compounds, we found a compound containing hydrazone and pyrazolo[3,4-d] pyrimidine structures (Fig. 2A and C). These two structures have been shown to possess enhancing activity of plant defense (Liu et al., 2014; Maher et al., 2022; Shi et al., 2021; Wang et al., 2024; Yu et al., 2022; Zhang et al., 2023a). The compound F1 formed strong hydrogen bonding interactions with the key residue MET-964, with a hydrogen bond length of less than 3.5 Å, which was an important contribution to the stabilization of pocket small molecules. In addition, due to the strong hydrophobicity of F1 backbone, it could form very good hydrophobic interactions with the amino acids VAL-902, GLY-967, VAL-894, LEU-1021, ALA-914, VAL-945, and played an important role in stabilizing the ligand in the active site of proteins (Fig. 2B and C).
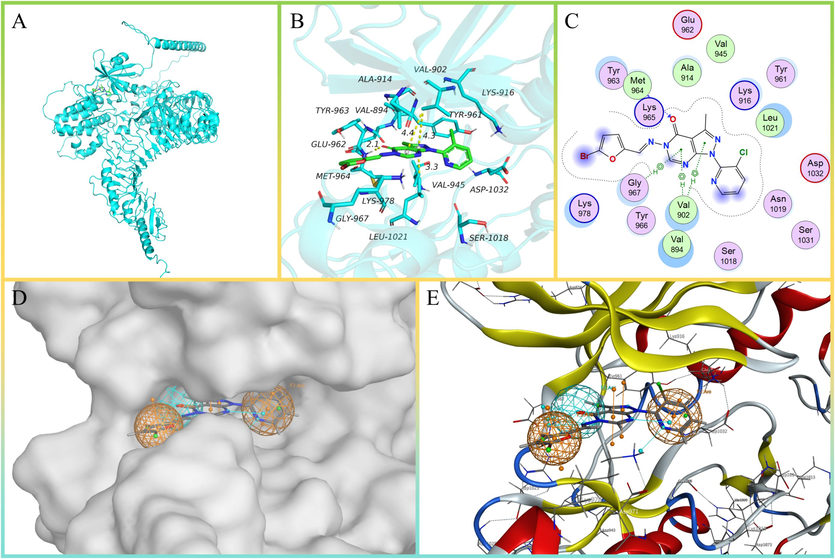
-
The docking model and pharmacophore model of SR160 with ligand. (A) The overall 3D structure of SR160-ligand complex. The backbone of protein was rendered in tube and colored in cyan. (B) A close view of the active site binding with ligand (green). Key residues interacted with ligand were rendered in stick and colored by cyan. (C) The 2D protein-F1 interaction diagram of SR160-ligand complex. Protein residues were rendered in circle and colored based on their properties: green, hydrophobic residue; purple, polar residue. (D) Docking-predicted binding packet of SR160 and ligand. (E) The pharmacophore model of SR160.
It was noteworthy that the hydrazone attached pyrazolo-pyrimidine faced inside the pocket, while the aromatic ring on the other side faced outside and the pocket space was open (Fig. 2B and C). Substitution in this aromatic ring region was considered and 44 additional compounds (F2-F45) were designed. Based on the interaction of F1 with SR160, the key binding characteristics were summarized as a pharmacophore model: hydrogen bonding of MET-964, hydrophobicity of the intermediate site, and hydrogen bonding receptor of ASP-1032 (Fig. 2D and E). Following that, 45 compounds (F1-F45) was screened by the pharmacophore model and all compounds scored below −8.0 (Supplementary Data.2).
3.2 Synthesis of target compounds
As shown in Fig. 3, ethyl 2-cyanoacetate A and triethyl orthoformate (triethyl orthoacetate) were added to a flask in a molar ratio of 1:1.2, with an appropriate amount of acetic anhydride used as a solvent. The mixture was refluxed and the reaction was monitored by TLC. After the completion of the reaction, the crude product was purified by column chromatography to give pure intermediate B. Subsequently, intermediate B was treated with 3-chlorodihydrazide pyridine in n-butanol under refluxing conditions to yield intermediate C (El-Tombary, 2013). Then the reaction of intermediate C with triethyl orthoformate gave D. Finally, intermediate E could easily be prepared by treating hydrazine hydrate (80 %) with D in ethanol at room temperature, yielding it in good yield (Y.Y. Wang et al., 2018). The title compounds were prepared by the reaction of intermediate E with different aldehydes in the presence of glacial acetic acid at reflux in ethanol with excellent yields.
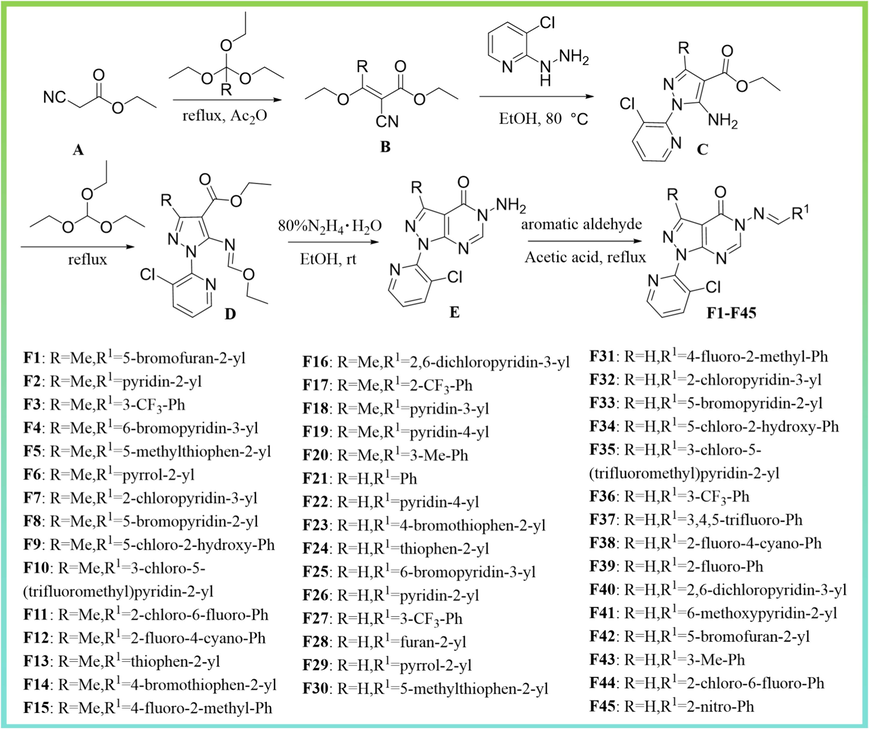
- Synthesis of pyrazolopyrimidine derivatives with hydrazone structure F1-F45.
3.3 Evaluation of biological activity
To verify the biological activity of the target compounds, a frictional TMV inoculation method was considered because systemins can improve plant defense against pathogenic microorganisms but plant resistance to TMV mainly depends on immune responses (Cirillo et al., 2022). This difference is advantageous in resolving whether the biological activity of compounds depends on the systemin pathway. The TMV treatment was performed after the compounds had been treated for 24 h to ensure that the plant was first stimulated by the compounds before being invaded by the virus. We used silicon carbide to rub on the leaves to provide the conditions for virus invasion. The effect of the defense response activated by F1-F45 was determined by investigating the incidence of the leaves. NNM is an antibiotic isolated from the Streptomyces noursei var-xichangensisn and is widely used to control crop viral diseases in China (Han et al., 2014). Therefore, NNM was used as a positive control to evaluate the activity of the compounds. Table 2 shows the results of antiviral activity for compounds F1 − F45 against CMV and TMV. Most of the compounds showed significant inhibitory activity against CMV. Compounds F2, F11, F17 and F30 showed 62.3 %, 66.6 %, 70.9 % and 62.7 % activity against CMV, respectively, with antiviral activity comparable to that of the commercial antiviral agent NNM (57.4 %). Meanwhile, the actions of compounds F19, F26, F37, and F38 against TMV were 78.4 %, 79.3 %, 77.0 %, and 81.0 %, respectively. Furthermore, the EC50 values was used to evaluate the effect of compounds on inducing plant resistance to TMV. The protective activities of compounds F19, F26, F37 and F38 were 78.8, 70.6, 76.5, and 74.6 μg/mL, respectively, which were slightly better than that of the control drug NNM (86.5 μg/mL) (Table 3).
The antiviral effect of the target compound was further confirmed through pot experiments. As shown in Fig. 4A and B, four days after TMV inoculation, only a small number of necrotic spots were observed on the right side of the leaves treated with the commercial antiviral agent NNM and compound F26. In contrast, a large number of necrotic spots appeared on the left side of the leaves that were not treated with the solution, and the leaves began to wither. As demonstrated in Fig. 4C and D, four days after CMV inoculation, only a few necrotic spots were observed on the right side of the leaves treated with the commercial antiviral agent NNM and compound F26, while a large number of necrotic spots appeared on the left side of the untreated leaves.
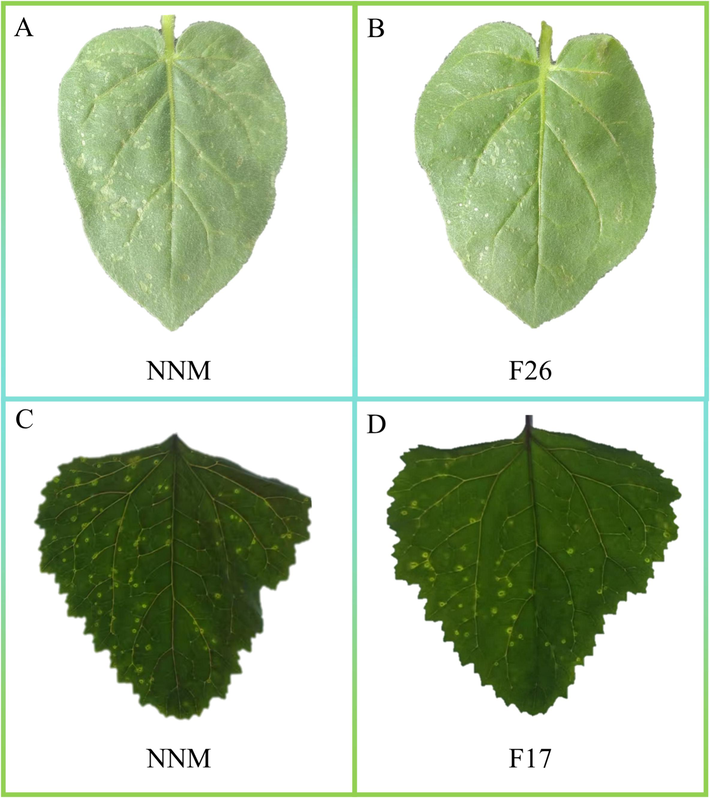
-
In vivo potted plant experiment. A: The in vivo inhibitory effect of commercial antiviral agent NNM on TMV at a concentration of 500 μg/mL. On the left is the control group CK, and on the right is the treatment group NNM. B: The in vivo inhibitory effect of compound F26 on TMV at a concentration of 500 μg/mL. On the left is the control group CK, and on the right is the treatment group F26. C: The in vivo inhibitory effect of commercial antiviral agent NNM on CMV at a concentration of 500 μg/mL. On the left is the control group CK, and on the right is the treatment group NNM. D: The in vivo inhibitory effect of compound F17 on CMV at a concentration of 500 μg/mL. On the left is the control group CK, and on the right is the treatment group NNM.
3.4 Plant response analysis
When plants suffer wounds, reactive oxygen species (ROS) accumulate rapidly because the membrane is damaged first. At the same time, the wound signal leads to the production of systemin and activates the systemin receptor. Subsequently, the MARK cascade signal is transmitted downward and leads to the corresponding defense response. The most responsive process in plants is oxidative stress, which is initiated when ROS accumulate and could also be induced by other defense pathways (Taiz et al., 2022). Activation of the systemin receptor leads to the production of defensive PIs, which is one of the typical defensive response features of wound signal activation (War et al., 2012). In addition, defensive secondary metabolism is also involved in the systemin-regulated defense response. Plants with systemin overexpression show obvious phenylpropanoids metabolic pathway, flavonoid synthesis and tyrosine metabolism (Pastor et al., 2018).
A frictional inoculation approach was used that made the first signal for plant was the wound. The enzyme activities of SOD and PPO were tested and the F26 treatment increased oxidative stress levels in plants (Fig. 5). From 3d to 7d, the PPO enzyme activities of the “F26 + TMV” group were 1.88, 1.43 and 1.03 times better than those of the blank control “CK + TMV”, respectively, and reached the peak on day 5 (Fig. 5B). Moreover, the enzyme activity of SOD for the “F26 + TMV”was also slightly higher than that of “CK + TMV” on days 3 and 5 (Fig. 5A).
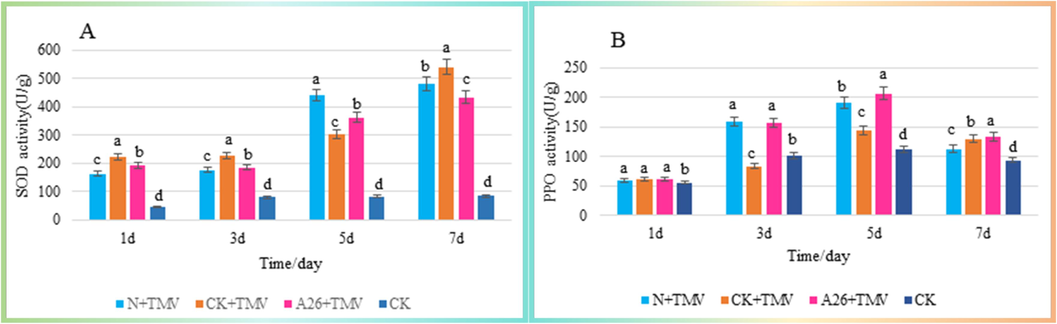
-
Results of defense enzyme activity. (A) SOD activity, (B) PPO activity. Straight sticks signify mean ± SD (n = 3). A one-way ANOVA followed by Duncan's test was used for significant differences at P < 0.05, marked with lowercase letters. Different lowercase letters indicate significant differences.
To further investigate the potential mechanism of F26, the total protein content of tobacco for “F26 + TMV” and “CK + TMV” were also studied by label-free LC-MS/MS. The proteomic data was divided into two parts. One part was differentially expressed proteins (DEPs) that the proteins were expressed in both groups, and the difference was whether the proteins expression of F26 was up-regulated or down-regulated compared with CK. The other part of the data was that some proteins were only expressed in F26 or CK during the detection range. This part was used for KEGG enrichment analysis.
As displayed in Fig. 6A, a total of 49 DEPs were identified, of which 17 were up-regulated, and 32 were down-regulated (fold change > 1.5, P < 0.05, red dots indicate up-regulated proteins, blue dots indicate down-regulated proteins).
Each protein involved in DEPs and KEGG was further analyzed in order to better understand the physiological changes in the tobacco. The proteomic data showed that Cysteine proteinase inhibitor (A0A059TCI3), Trypsin inhibitor (Q7M1P5) and NtLTP4 triggered by wound signals were expressed in F26 treatment, while immune react-related disease resistance proteins (Q5DJS5, Q6LBM4, P93362) were more expressed in CK. Both Trypsin inhibitor and Cysteine proteinase inhibitor belong to PIs and are involved in plant defense responses to wound, insect attack and pathogen invasion (Botelho et al., 2008; Jongsma et al., 1994; Pearce et al., 1993; Rustgi et al., 2017). NtLTP4 is a lipid transfer protein that can interact with WIPK (wound-induced protein kinase) and up-regulate the expression of defense-related genes to improve the resistance to pathogens (Xu et al., 2022). This implies that F26 could take the edge off the harm of TMV to tobacco by enhancing the defensive response induced by wound signals. The phenylpropanoids metabolic pathway, flavonoid synthesis and tyrosine metabolism were also affected by F26 treatment (Fig. 6B). These features are similar to systemin-induced defense responses. Overexpression of systemin in plants induces accumulation of lignans that are dehydrodimers of monolignols, but the precursor compounds of phenylpropanoid such as caffeic and ferulic acids are reduced (Pastor et al., 2018). F26 treatment had more monolignols because the expression of peroxidase (A0A1S3ZDR0, A0A1S4A886, A0A1S4A3W2, A0A1S3Y048, Q50LG5, Q9XIV9) catalyzing the conversion of monolignol to lignin. This may contribute to the accumulation of lignans. The expression of HQT (Hydroxycinnamoyl CoA quinate transferase, Q70G33) and 4CL (4-coumarate--CoA ligase, A0A1S3ZTJ1) were up-regulated in F26 treatment. HQT is a key enzyme in the synthesis of chlorogenic acid which is an antioxidant in plants and has a wide range of pharmacological activities, such as antibacterial, cardioprotective, antiviral, hepatoprotective and so on (Naveed et al., 2018). Overexpression of HQT caused tomato to accumulate more chlorogenic acid and improved antioxidant capacity and resistance to bacterial infection (Niggeweg et al., 2004). 4CL catalyzes p-coumarate to p-coumaroyl-CoA in the phenylpropanoid pathway (Wagner et al., 2012). The eight phenylpropanoid pathway proteins were further validated by RT-qPCR experiments. The results (Fig. 6C) revealed that compound F26 could forcefully activate the expression levels of genes NtLTP4, Q70G33, and A0A1S3ZTJ1, and slightly downregulate the expression levels of genes A0A1S3ZDR0, A0A1S4A886, A0A1S4A3W2, A0A1S3Y048, Q50LG5, and Q9XIV9. These differences in phenylpropanoid synthesis pathways may be caused by the different experimental conditions, since the systemin overexpressed in the work of Victoria Pastor et al. were under normal growth conditions, while our work was performed under viral infection. In addition, photosynthetic proteins and energy metabolism proteins were also highly expressed in F26 treatment, but this response may be post-reactive because F26 treatment had better leaf integrity than CK.
![Analysis of plant response after F26 treatment. (A) Volcano plot of the relative protein abundance changes between the compound F26 + TMV and CK + TMV treatments. The red dots represent significantly up-regulated proteins, whereas the blue dots represent significantly down-regulated proteins, and the grey spots represent unchanged proteins in both treatment groups. (B) Enrichment map of KEGG pathway of differentially expressed proteins in tobacco treated with the compound F26. (C) Gene expression analysis of the related genes of the phenylpropanoid biosynthesis pathway by qRT-PCR. (β-actin gene served as the internal control). [mean values displayed in each bar followed by different letters significantly differ according to Student's t-test (**P < 0.005, ****P < 0.0001). Vertical bars indicate SD (n = 3)].](/content/184/2024/17/8/img/10.1016_j.arabjc.2024.105849-fig6.png)
-
Analysis of plant response after F26 treatment. (A) Volcano plot of the relative protein abundance changes between the compound F26 + TMV and CK + TMV treatments. The red dots represent significantly up-regulated proteins, whereas the blue dots represent significantly down-regulated proteins, and the grey spots represent unchanged proteins in both treatment groups. (B) Enrichment map of KEGG pathway of differentially expressed proteins in tobacco treated with the compound F26. (C) Gene expression analysis of the related genes of the phenylpropanoid biosynthesis pathway by qRT-PCR. (β-actin gene served as the internal control). [mean values displayed in each bar followed by different letters significantly differ according to Student's t-test (**P < 0.005, ****P < 0.0001). Vertical bars indicate SD (n = 3)].
4 Conclusion
In this study, based on the conserved characterization of the kinase domains of the Leucine-rich repeat protein kinase family, we constructed systemin receptors as a virtual screening model for drug discovery. A potentially active structure containing hydrazone and pyrazolo[3,4-d] pyrimidine was obtained by virtual screening. Then, we designed a series of compounds containing this structure and evaluated their ability to bind SR160 by molecular docking. Based on the results, the biological activities of the designed compounds were evaluated by frictional inoculation of the virus. Compound F26 exhibited excellent antiviral activity with an EC50 value of 70.6 μg/mL against TMV. Furthermore, the physiological changes of plants after compounds treatment were analyzed by proteomic data. Compound F26 enhanced wound signaling-induced expression of protease inhibitors and significantly enhanced the phenylpropionate metabolic pathway and flavonoid pathway. These changes were similar to those in overexpressed systemin plants. In conclusion, the present study demonstrated that designed drug based on the systemin receptor was a viable option and that F26 enhanced plant defense against pathogenic microorganisms through the wound signaling pathway. To the best of our knowledge, this is the first time that the systemin receptor has been used as a potential target protein for the development of drug with plant activity.
CRediT authorship contribution statement
Ya Wang: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. Zhichao Zhao: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. Renjiang Guo: Investigation. Yao Tang: Investigation. Shengxin Guo: Formal analysis. Ying Xu: Validation. Wei Sun: Validation. Hong Tu: Formal analysis. Jian Wu: Writing – review & editing, Methodology, Conceptualization.
Acknowledgement
This work was supported by the National Key R&D Program of China (No. 2023YFD1700602), the Science and Technology Plan Project of Guizhou Province (Qiankehezhicheng[2024] the general 083), the Program of Introducing Talents to Chinese Universities (No. D20023), the Central Government Guides Local Science and Technology Development Fund Projects (Qiankehezhongyindi (2023) 001).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- An overview of plant defense-related enzymes responses to biotic stresses. Plant Gene. 2021;27:100302
- [CrossRef] [Google Scholar]
- Trypsin inhibitors in passion fruit (Passiflora f. edulis flavicarpa) leaves: accumulation in response to methyl jasmonate, mechanical wounding, and herbivory. J. Agric. Food Chem.. 2008;56:9404-9409.
- [CrossRef] [Google Scholar]
- Signal transduction in the wound response of tomato plants. Philos. Trans. r. Soc. Lond. B Biol. Sci.. 1998;353:1495-1510.
- [CrossRef] [Google Scholar]
- Plant leucine-rich repeat receptor kinase (LRR-RK): Structure, ligand perception, and activation mechanism. Molecules. 2019;24
- [CrossRef] [Google Scholar]
- Systemin peptide application improves tomato salt stress tolerance and reveals common adaptation mechanisms to biotic and abiotic stress in plants. Environ. Exp. Bot.. 2022;199:104865
- [CrossRef] [Google Scholar]
- The emerging evidence of the parkinson pandemic. J. Parkinsons Dis.. 2018;8:S3-S8.
- [CrossRef] [Google Scholar]
- Synthesis, anti-inflammatory, and ulcerogenicity studies of novel substituted and fused pyrazolo[3,4-d]pyrimidin-4-ones. Sci. Pharm.. 2013;81:393-422.
- [CrossRef] [Google Scholar]
- Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6:751-759.
- [CrossRef] [Google Scholar]
- Inspired by nature: Isostere concepts in plant hormone chemistry. J. Agric. Food Chem.. 2023;71:18141-18168.
- [CrossRef] [Google Scholar]
- Origin and evolution of the plant immune system. New Phytol.. 2019;222:70-83.
- [CrossRef] [Google Scholar]
- Induction of systemic resistance against tobacco mosaic virus by Ningnanmycin in tobacco. Pestic. Biochem. Physiol.. 2014;111:14-18.
- [CrossRef] [Google Scholar]
- Computer aided drug design: Success and limitations. Curr. Pharm. Des.. 2016;22:572-581.
- [CrossRef] [Google Scholar]
- Peptide-based protease inhibitors from plants. Drug Discov. Today. 2019;24:1877-1889.
- [CrossRef] [Google Scholar]
- Plant protection against viruses: an integrated review of plant immunity agents. Int. J. Mol. Sci.. 2023;24:4453.
- [CrossRef] [Google Scholar]
- Trypsin inhibitor activity in mature tobacco and tomato plants is mainly induced locally in response to insect attack, wounding and virus infection. Planta. 1994;195:29-35.
- [CrossRef] [Google Scholar]
- Virtual screening-based drug development for the treatment of nervous system diseases. Curr. Neuropharmacol.. 2023;21:2447-2464.
- [CrossRef] [Google Scholar]
- Insights into a class of natural eugenol and its optimized derivatives as potential tobacco mosaic virus helicase inhibitors by structure-based virtual screening. Int. J. Biol. Macromol.. 2023;248:125892
- [CrossRef] [Google Scholar]
- Design, synthesis, and antiviral, fungicidal, and insecticidal activities of tetrahydro-β-carboline-3-carbohydrazide derivatives. J. Agric. Food Chem.. 2014;62:9987-9999.
- [CrossRef] [Google Scholar]
- Role of computer-aided drug design in modern drug discovery. Arch. Pharm. Res.. 2015;38:1686-1701.
- [CrossRef] [Google Scholar]
- Recent green approaches for the synthesis of pyrazolo[3,4-d]pyrimidines: A mini review. Arch Pharm (weinheim). 2022;355:2100470.
- [CrossRef] [Google Scholar]
- The tomato brassinosteroid receptor BRI1 increases binding of systemin to tobacco plasma membranes, but is not involved in systemin signaling. Plant Mol. Biol.. 2009;70:603-616.
- [CrossRef] [Google Scholar]
- Colonization of solanum melongena and vitis vinifera plants by botrytis cinerea is strongly reduced by the exogenous application of tomato systemin. J Fungi (basel). 2020;7:1-15.
- [CrossRef] [Google Scholar]
- Chlorogenic acid (CGA): A pharmacological review and call for further research. Biomed. Pharmacother.. 2018;97:67-74.
- [CrossRef] [Google Scholar]
- Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol.. 2004;22:746-754.
- [CrossRef] [Google Scholar]
- Accurate and easy method for systemin quantification and examining metabolic changes under different endogenous levels. Plant Methods. 2018;14
- [CrossRef] [Google Scholar]
- Tomato Systemin induces resistance against Plectosphaerella cucumerina in Arabidopsis through the induction of phenolic compounds and priming of tryptophan derivatives. Plant Sci.. 2022;321:111321
- [CrossRef] [Google Scholar]
- Systemin, hydroxyproline-rich systemin and the induction of protease inhibitors. Curr. Protein Pept. Sci.. 2011;12:399-408.
- [CrossRef] [Google Scholar]
- Purification and characterization from tobacco (Nicotiana tabacum) leaves of six small, wound-inducible, proteinase isoinhibitors of the potato inhibitor II family. Plant Physiol.. 1993;102:639-644.
- [CrossRef] [Google Scholar]
- The complex world of plant protease inhibitors: Insights into a Kunitz-type cysteine protease inhibitor of Arabidopsis thaliana. Commun. Integr. Biol.. 2017;11
- [CrossRef] [Google Scholar]
- Jasmonate signaling in plant development and defense response to multiple (a)biotic stresses. Plant Cell Rep.. 2013;32(7):1085-1098.
- [CrossRef] [Google Scholar]
- The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. PNAS. 2002;99:9585-9590.
- [CrossRef] [Google Scholar]
- Systemic signaling in the wound response. Curr. Opin. Plant Biol.. 2005;8:369-377.
- [CrossRef] [Google Scholar]
- Floro-pyrazolo[3,4-d]pyrimidine derivative as a novel plant activator induces two-pathway immune system. Phytochemistry. 2021;184
- [CrossRef] [Google Scholar]
- MYC5 is involved in jasmonate-regulated plant growth, leaf senescence and defense responses. Plant Cell Physiol.. 2017;58:1752-1763.
- [CrossRef] [Google Scholar]
- Synthesis and antiviral activity of novel chiral cyanoacrylate derivatives. J. Agric. Food Chem.. 2005;53:7886-7891.
- [CrossRef] [Google Scholar]
- Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res.. 2013;23(11):1326-1329.
- [CrossRef] [Google Scholar]
- Systemin/jasmonate-mediated systemic defense signaling in tomato. Mol. Plant. 2011;4:607-615.
- [CrossRef] [Google Scholar]
- Taiz, L., Møller, I.M. (Ian M., Murphy, A.S., Zeiger, E., 2022. Plant physiology and development.
- Lignification and lignin manipulations in conifers. Adv. Bot. Res.. 2012;61:37-76.
- [CrossRef] [Google Scholar]
- The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat. Plants. 2018;4:152-156.
- [CrossRef] [Google Scholar]
- Hydrazone derivatives in agrochemical discovery and development. Chin. Chem. Lett.. 2024;35:108207
- [CrossRef] [Google Scholar]
- Pyrazolo[3,4-d]pyrimidine derivatives containing a Schiff base moiety as potential antiviral agents. Bioorg. Med. Chem. Lett.. 2018;28:2979-2984.
- [CrossRef] [Google Scholar]
- Synthesis and antiviral bioactivity of novel chalcone derivatives containing purine moiety. Chin. Chem. Lett.. 2018;29:127-130.
- [CrossRef] [Google Scholar]
- Mechanisms of plant defense against insect herbivores. Plant Signal. Behav.. 2012;7:1306-1320.
- [CrossRef] [Google Scholar]
- Design, synthesis, antiviral bioactivity and three-dimensional quantitative structure-activity relationship study of novel ferulic acid ester derivatives containing quinazoline moiety. Pest Manag. Sci.. 2017;73:2079-2089.
- [CrossRef] [Google Scholar]
- Synthesis and antiviral activities of novel 1,4-pentadien-3-one derivatives bearing an emodin moiety. Chin. Chem. Lett.. 2016;27:948-952.
- [CrossRef] [Google Scholar]
- WIPK-NtLTP4 pathway confers resistance to Ralstonia solanacearum in tobacco. Plant Cell Rep.. 2022;41:249-261.
- [CrossRef] [Google Scholar]
- The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220-2236.
- [CrossRef] [Google Scholar]
- Design, synthesis, and bioactivity of spiro derivatives containing a pyridine moiety. J. Agric. Food Chem.. 2022;70:15726-15736.
- [CrossRef] [Google Scholar]
- Novel trifluoromethylpyridine piperazine derivatives as potential plant activators. Front. Plant Sci.. 2022;13:1086057.
- [CrossRef] [Google Scholar]
- Trifluoromethylpyridine piperazine derivatives: synthesis and anti-plant virus activity. Pest Manag. Sci.. 2023;79:2571-2580.
- [CrossRef] [Google Scholar]
- Piperazine: Its role in the discovery of pesticides. Chin. Chem. Lett.. 2023;34:108123
- [CrossRef] [Google Scholar]
- the spread and transmission of sweet potato virus disease (SPVD) and its effect on the gene expression profile in sweet potato. Plants (basel). 2020;9
- [CrossRef] [Google Scholar]
- Expression of tomato prosystemin gene in Arabidopsis reveals systemic translocation of its mRNA and confers necrotrophic fungal resistance. New Phytol.. 2018;217:799-812.
- [CrossRef] [Google Scholar]
- Systemin-mediated long-distance systemic defense responses. New Phytol.. 2020;226:1573-1582.
- [CrossRef] [Google Scholar]
- The proteomics and metabolomics studies of GZU001 on promoting the Merisis of maize (Zea mays L.) roots. BMC Plant Biol.. 2023;23
- [CrossRef] [Google Scholar]
- Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM. Cell Discov. 2024;10
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2024.105849.
Appendix A
Supplementary data
The following are the Supplementary data to this article:
Supplementary Data 1
Supplementary Data 1
Supplementary Data 2
Supplementary Data 2
Supplementary Data 3
Supplementary Data 3
Supplementary Data 4
Supplementary Data 4







