Translate this page into:
Mixed dye degradation by Bacillus pseudomycoides and Acinetobacter haemolyticus isolated from industrial effluents: A combined affirmation with wetlab and in silico studies
⁎Corresponding author. shu_genetics@yahoo.com (Sudhangshu Kumar Biswas)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The environment-friendly bacterial strains are used for wastewater treatment due to their high degrading capability and cost-effectiveness. In the present study, we isolated, identified, characterized, and optimized culture condition ( Temperature 35 °C for both, time up to 96 h, pH 7 and 7.5 respectively) of two dye degrading bacteria from industrial effluents. Morphological, biochemical, and molecular identification confirmed those strains as Bacillus pseudomycoides and Acinetobacter haemolyticus. Spectrophotometric methods were used to investigate the dye degradation(single and mixed dye) capability of these two bacteria. These strains were the potential for degrading methylene green (MG), basic violet (BV) and acid blue (AB) dyes. In case of MG + BV, B. pseudomycoides, and A. haemolyticus showed a degradation rate of 74% and 75%, respectively. While degradation was found 75% and 82% for MG + AB combination, 73% and 73% for AB + BV combination, and 80% and 82% for MG + BV + AB combination respectively. Azoreductase enzymes from bacteria are essential for breaking down the azo bond in textile azo dyes. In molecular docking, the binding energy of three docking complexes (protein and MG, protein and BV, protein and AB) were −6.3, −6.6, and −6.8 Kcal/mol, respectively. The binding stability of the docked complexes was ensured by the root mean square deviations (RMSD), solvent accessible surface area (SASA), radius of gyration (Rg) and hydrogen bond in a molecular dynamics simulation study, indicating strong and stable binding. This study revealed that both B. pseudomycoides and A. haemolyticus could decolorize single and mixed dyes efficiently. As a result, both the strains could be used in further research to apply their potentiality in large-scale dye degradation in the future.
Keywords
Textile effluents
Mixed dye degradation
16s rDNA sequencing
Molecular docking
1 Introduction
Environmental pollution is the world’s most serious socio-economic and health issue, with water pollution being the most pressing concern today (Deng et al., 2020). The widespread use of dyes contributes to the environmental challenges associated with textile operations (Zou et al., 2020). Bangladesh is now the second-largest garment exporter, and its economy is mostly based on textile industries (Agarwal et al., 2017). Synthetic dye is widely used in textile printing and dyeing, as well as cosmetics, plastics, photography, and paper industries (Leulescu et al., 2018; Manzoor and Sharma, 2019; Swarnkumar Reddy and Osborne, 2020). The dyes are then released into sewers and drains, which run into the natural water resources such as rivers and other water sources, affecting water diversity (Islam et al., 2011; Joshi et al., 2021; Kanu et al., 2011). More than 10,000 distinct dyes and pigments are utilized in industry, and over 7 × 105 tons of synthetic dyes are manufactured globally each year (Farhan et. al., 2019; Gürses et al., 2016; Javaid et al., 2021). The most commonly used dyes in the textile industry are methylene green, acid blue, and basic violet. Methylene green is a heterocyclic aromatic chemical compound. The chemical formula of Methylene green is C16H17ClN4O2S and the IUPAC name [7-(dimethylamino)-4-nitrophenothiazin-3-ylidene]-dimethylazanium chloride, with a molecular weight of 364.85 g/mol and a maximum light wavelength of 614 nm. Acid blue is an acid dye with water soluble anionic, has the chemical formula C20H13N2NaO5S and the IUPAC name Sodium 1-amino-4-anilino-9,10-dioxoanthracene-2-sulfonate, with a molecular weight of 416.38 g/mol and a maximum wavelength of 620 nm. Likewise, basic violet is a basic dye has the chemical formula C30H35ClN2O3 and the IUPAC designation [6-(diethylamino)-9-(2-ethoxycarbonylphenyl) xanthen-3-ylidene]. -diethylazanium; chloride has a molecular weight of 507.1 g/mol and a wavelength of 590 nm.(See Table 1.).
Complex
Amino Acid Residues
Bond Type
Distance(Å)
Docking Energy (Kcal/mol)
Azoreductase-Methylene Green
Pro101
A
4.50
−6.3
His10
PA
5.30
Trp103
PA
4.78
Azoreductase-Basic violet
Asp116
H
3.38
−6.6
Lys112
H
3.65
Asp116
PA
3.53
Trp60
PPT
5.32
Phe172
PPT
4.84
Ile52
A
3.68
Ile168
A
4.66
Phe57
PA
4.70
Ala119
PA
4.55
Ilu169
PA
5.38
Azoreductase- Acid Blue
Asn104
H
2.24
−6.8
His186
PS
4.93
Pro11
A
3.79
His10
PA
4.28
Phe18
PA
5.22
Every year up to 200,000 tons of these dyes are mixed with effluents in the textile industry during dyeing and finishing operations due to the inefficiencies in the dyeing process (Wan et al., 2021). These dyes are unreacted up to 50% when released into effluents and difficult to remove by conventional wastewater treatment due to high water solubility (Moreira et al., 2004; Ogola et al., 2015; Rodríguez Couto, 2009). These effluents are harmful to humans, plants, and living organisms (Mahbub et al., 2012). To reduce dye toxicity from textile effluents, industrial effluents containing azo dyes must be processed before discharge into the environment (Bharagava and Chowdhary, 2018). For the treatment of textile wastewater, distinct physical, chemical, and biological methods such as adsorption, chemical precipitation, photolysis, chemical oxidation, and reduction are used (Verma, 2021). These methods are inefficient, expensive, and have limited applicability, as well as producing trash that is is difficult to dispose of (Dahiya and Nigam, 2020). So, in order to reduce and eliminate synthetic dyes within few a hours, environmentally friendly, cost-effective, and new strategies are required (Fletcher et al., 2021; Lie et al., 1996). Bioremediation is non-hazardous, cost-efficient, environmentally friendly, or an even more effective alternative to traditional methods for the treatment of textile waste (Fletcher et al., 2021). Bioremediation employing microbes such as bacteria, fungi, algae, yeast, and mixed culture is a promising technique (dos Santos et al., 2007; Jørgensen et al., 2017; Li et al., 2019; Huda et. al., 2021). The bacteria's ability to adapt and degrade textile effluents at high concentrations offers them an edge in the treatment of textile effluents. The employment of bacterial techniques to degrade synthetic dyes, notably azo dyes, may be advantageous (Mishra et al., 2020). Therefore, isolation, identification, and preservation of different dye degrading bacteria is an essential part of research and development.
Various oxidoreductive enzymes like peroxidases, laccases, polyphenol oxidases, and azoreductases have been explored for decolorization and degradation of azo dyes (Srinivasan et al., 2017). So, molecular docking and simulation is an inexpensive method for optimizing the significance and interaction among protein-ligands. This combinatorial approach of employing bioinformatics tools followed by wet-lab analysis can be an effective way to screen for possible dye decolorizing bacterial systems that can then be used in real-time wastewater treatment in the textile industries. In the present study, Bacillus pseudomycoides and Acinetobacter haemolyticus, which degrade mixed dyes, were isolated, identified, and optimized from textile effluents. Then, employing azoreductase enzymes from Bacillus spp., bioremediation of three textile azo dyes, methylene green, acid blue, and basic violet, was explored.
Present study focuses on both single and mixed dye degradation capability of the isolated bacteria while most of the previous researcher investigated only the single dye degradation. Furthermore, in silico binding interaction and stability of the two newly isolated bacterial strains is also investigated.
2 Materials and methods
2.1 Sample collection and screening of dye degrading bacteria
Textile effluents were collected aseptically in a sterile plastic bottle from Rana Textile, Kushtia, Bangladesh, and transported to the Laboratory right away. The bacterial strains were screened using the Paul et al., (2020) and Schoenborn et al., (2004) methods. The collected effluents were diluted serially to obtain a dilution factor of 10−4, 10−5, 10−6, 10−7, and 0.1 ml of the diluted samples were spread onto LB agar (Oxoid-UK) plates and incubated overnight (16 h) at 35 °C, pH-7 (Abbas et al., 2014). The chemicals and materials used in this research were analytical grade and procured following the rules and regulation of Islamic University, Kushtia-7003, Bangladesh and the purities of all dyes were confirmed before purchased.
2.2 Single dye degradation assay
Each bacterial colony from spread cultured plate was inoculated in one particular dye supplemented with LB agar plate through spot culture technique. Firstly, eight dye degrading bacterial isolates were selected by looking the clear zone around the spot culture. After preliminary selection of dye degrading bacteria through dye supplemented agar media the dye degradation was observed in dye supplemented LB liquid media. The dye degradation rate of eight selected strains was measured at 614 nm, 620 nm, and 590 nm for methylene green (MG), acid blue (AB), and basic violet (BV) dye, respectively, according to (Liu et al., 2021). Briefly, the concentration of different dyes was adjusted to 100 mg/l using LB liquid media (Oxoid-UK) in a serum bottle where 100 µL of each bacterial strain was added. Furthermore, the concentration of cells/biomass of bacterial culture ranged from 1.7 × 107 CFU/ml to 3.8 × 107 CFU/ml. In this research, each experiment was performed in triplicate. The optical density was observed every 12 h for the first 96 h where the culture bottles were incubated at 35 °C. The optical density was measured by using a spectrophotometer (Analytic Gena, Germany), and the dye degradation rate was calculated using the following formula (Agrawal et al., 2014).
For mixed dye degradation, the concentrations were also adjusted to 100 ppm by using an equal amount of methylene green, acid blue, and basic violet dyes into the LB medium. Then, 100 µL of the best two dye degrading isolates (S-11 and S-15) were separately suspended into dyes containing serum bottles. The culture bottles were kept at 35 °C for 96 h, and optical density was measured every 12 h. The optical density of different single dyes was measured at their average maximum wavelength. In supplementary Fig. 7 we have showed mechanisms of mixed dye degradation schematically.
2.3 Growth parameters optimization of isolated bacteria
Different growth parameters including pH, temperature, carbon sources, nitrogen sources, and percentage of salt were optimized according to Pramanik et al., (2021). The pH gradients were set to 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, where the temperature were set to 25 °C, 30 °C, 35 °C, 40 °C and 45 °C. The carbon sources were sucrose (Innovating Science- USA), glucose (Innovating Science- USA), dextrose (Innovating Science- USA), glycerol (Innovating Science- USA), the nitrogen sources were peptone (Oxoid-UK), yeast extract (Oxoid-UK), urea and tryptone (Oxoid-UK), and NaCl (Chemall-India) and the percentage were set to 0.5%, 1%, 2%, 3 %, 5%, and 10%, respectively in LB medium. Data were recorded at 12 h intervals from 0 h to 72 h.
2.4 Morphological and biochemical characters observation
Different morphological tests such as Gram staining and motility and biochemical tests like methyl red (Somatco-Saudi) test, catalase test, macConkey (HiMedia-India) test, mannitol salt (Oxoid-UK) test, urea hydrolysis (Urease-HiMedia-India) test, starch (Oxoid-UK) hydrolysis test, triple sugar iron (HiMedia-India) test, simmon citrate (Merck, India) test, bismuth sulfate agar (HiMedia-India) test, eosin methylene blue agar (Thermo Fisher Scientific-USA) test, and oxidase test were carried out according to Usta and Demirkan (2018).
2.5 Sensitivity test to antibiotics
A total of 12 (twelve) antibiotics namely kanamycin (30 µg), tetracycline (30 µg), ampicillin (10 µg), amoxicillin (30 µg), ciprofloxacin (5 µg), penicillin-G (10 µg), cefuroxime (30 µg), doxycycline (30 µg), gentamycin (10 µg), erythromycin (15 µg), cefixime (5 µg), and ceftazidime (30 µg) were selected for antibiotic sensitivity test of selected strains. The antibiotic sensitivity of isolated strains was performed according to Paul et al. (2020).
2.6 Molecular identification of the selected strains
Molecular identification of the strains was carried out according to the method described by Stach et al., (2001). Briefly, the PCR mixture contains 10 µL Hot Start Green Master Mix, 1 µL of genomic DNA, 1 µL of each primer, and 7 µL of ddH2O. The universal forward (27F: 5-AGA GTT TGA TCC TGG CTC AG-3) and reverse (1492R: 5- CGG TTA CCT TGT TAC GAC TT −3) primers were used for the PCR amplification of DNA. The PCR conditions were as follows: initial denaturation at 95 °C for 3 min, denaturation at 95 °C for 30 sec, primer annealing at 48 °C for 30 sec, extension at 72 °C for 90 sec and final extension at 72 °C for 5 min. The PCR products were purified and both strands were sequenced by a genetic analyzer (Prism 310, USA) and the sequences were edited using bioinformatics software Chromas. Phylogenetic trees were constructed using MEGA X software and the basic local alignment search tool (BLAST) was performed through the NCBI database ( https://www.blast.ncbi.nlm.nih.gov/Blast.cgi ).
2.7 Ligand preparation
The three-dimensional structure of methylene green, acid violet, and acid greens were retrieved from the PubChem database (Kim et al., 2016). The energy minimization of the ligand molecules was performed by the Avogadro software package by employing the mmff94 force field (Halgren, 1996a, 1996b).
2.8 Protein preparation
The three-dimensional structure of Azoreductase from Bacillus sp. (PDB ID:6QU0) was retrieved from the Protein Data Bank (Berman et al., 2002, , 2000). The water and heteroatoms were removed from the structure and cleaned in Pymol (Mooers, 2020). The cleaned structure was subjected to energy minimization in YASARA (Land and Humble, 2018) using the AMBER14 force field (Case et al., 2014).
2.9 Molecular docking
The docking study of the target protein and ligand structure was performed in the Auto Dock Vina software package (Jaghoori et al., 2016; Trott and Olson, 2009; Vieira and Sousa, 2019). The ligand molecules were converted to PDBQT format, which is an Auto Dock Vina-acceptable format. Therefore, the box size and grid box center were set to (X:-30.34, Y:-24.18, Z: 9.92), (X: 45.53, Y: 65.82, Z: 38.26), respectively. Finally, the binding interactions were analyzed using Pymol (Mooers, 2020) and the Discovery Studio software package (BIOVIA, 2016).
2.10 Molecular dynamics
The molecular dynamics simulation was conducted in the YASARA dynamics (Land and Humble, 2018) software package using the AMBER14 force field (Wang et al., 2004). The complex structure was initially cleaned, optimized and hydrogen bond network systems were oriented (Krieger et al., 2012). The cubic simulation cell with periodic boundary conditions was created using the TIP3P solvation model (Harrach and Drossel, 2014). The simulations cells were extended by 20 Å at each side of the complexes. The physiological conditions were set to 298 K, pH 7.4, and 0.9% NaCl (Krieger and Vriend, 2015). The initial energy minimizations of the simulation cell were achieved by steepest gradient algorithms by the simulated annealing method (5000 cycles). The time step of the simulations systems was set to 2.0 fs. The long-range electrostatic interactions were calculated by the Particle Mesh Ewald (PME) algorithms by a cut-off radius of 8.0 Å (Essmann et al., 1995; Harvey and De Fabritiis, 2009; Krieger et al., 2006). The simulation trajectories were saved after every 100 ps interval. By following constant pressure and Berendsen thermostat, the simulation was extended for 100 ns, and trajectories were used to analyze the root mean square deviations (RMSD), hydrogen bond, solvent accessible surface area (SASA), and radius of gyration (Rg) (Dutta et al., 2021; Mahmud et al., 2021a; Mahmud, et al., 2021b; Mahmud et al., 2021c; Mousavi et al., 2021).
2.11 Statistical analysis
Duncan’s multiple range test (DMRT) was used to analyze the significance of each group data at a p < 0.05 level of significance in a one-way analysis of variance in SPSS Statistics 26 software. Graph Pad Prism 8.0.2.263 was used for preparing all figures.
3 Results
3.1 Screening of dye degrading bacteria
Among the 22 (twenty-two) isolated strains, only 8 (eight) strains were capable of dye degradation (supplementary figure-1). These strains were designated as S-1, S-2, S-3, S-5, S-9, S-11, S-15, and S-17.
3.2 Single dye degradation
The dye degradation rates by the selected eight bacterial strains are shown in supplementary table-1 and from the table it is seen that the selected strains, S-1, S-2, S-3, S-5, S-9, S-11, S-15 and S-17 had a degradation rate of 81%, 79%, 84%, 89%, 89%, 94%, 91%, and 88%, respectively (Fig. 1). S-11 and S-15 were chosen for further investigations due to their significant degradation capability where the S-11 was the best for MG, AB and BV dye degrading bacteria (Fig. 1-f) among the isolates in this study.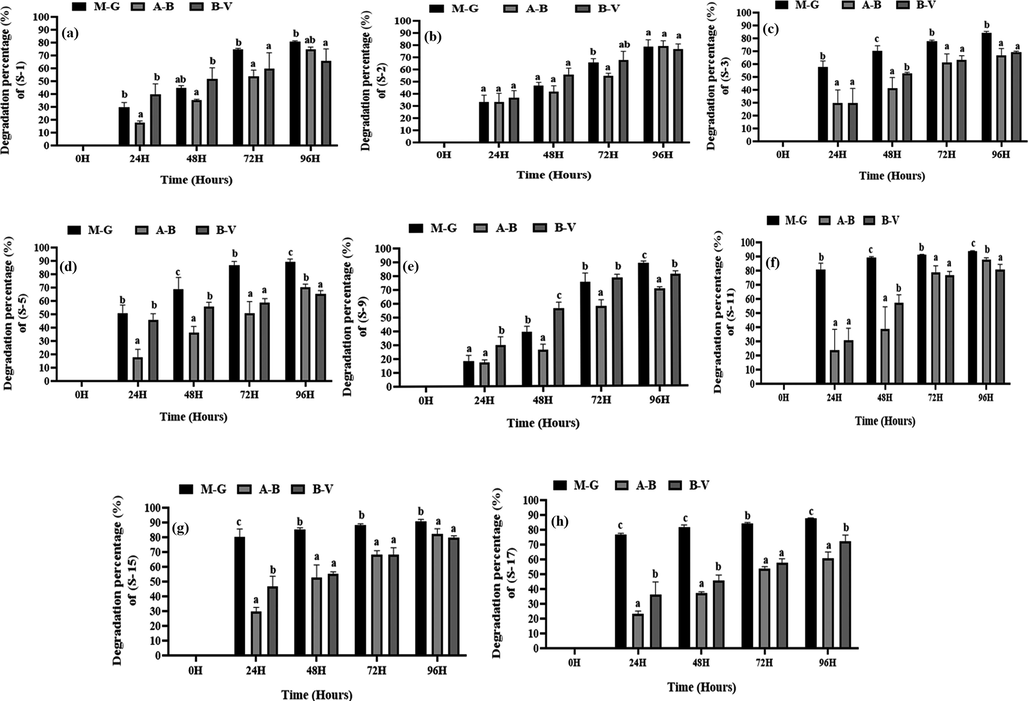
Single dye degradation rates of different isolated strains. Here (a), (b), (c), (d), (e), (f), (g), (h) indicate the degradation rate of S-1, S-2, S-3, S-5, S-9, S-11, S-15, and S-17, respectively. MG, AB, and BV indicate methylene green, acid blue, and basic violet dye, respectively. Different letters indicate the significance differences at a p < 0.05 significance level.
3.3 Bacterial growth optimizations
The optimum pH for S-11 and S-15 growth was 7.0 and 7.5, respectively, while the optimum temperature was 35 °C for both of the isolates (Fig. 2). In terms of carbon sources, S-11 performed best on dextrose, and S-15 performed best on glucose (Fig. 3). Both the strains performed best when yeast extract was used as a nitrogen source. (Fig. 3). S-11exhibitted better performance at 0.5% NaCl concentration and S-15 was best at 1.0% concentration (Fig. 3).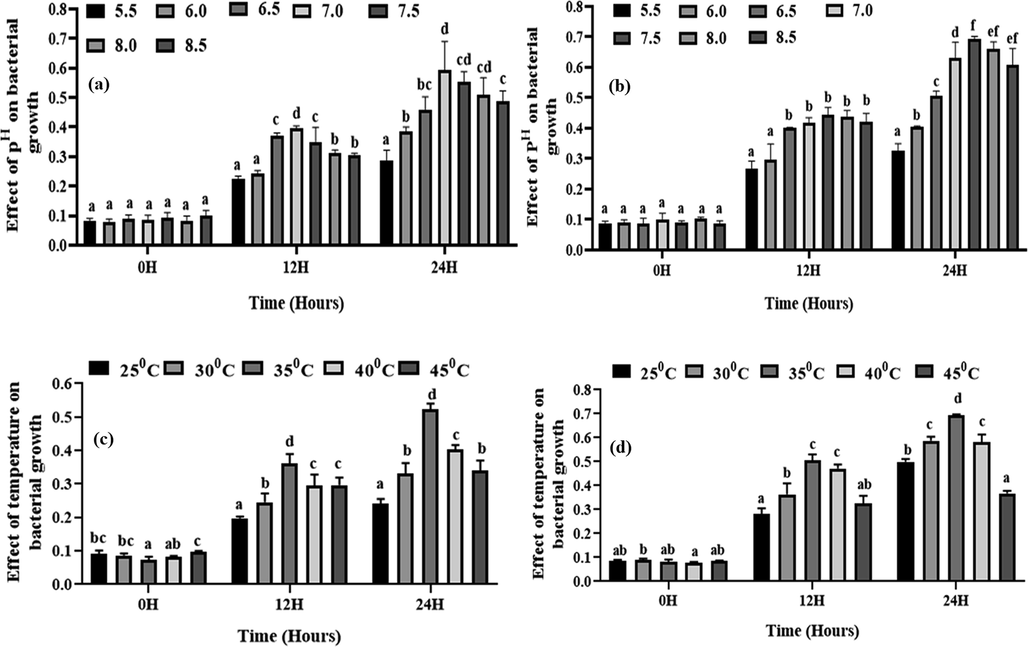
Effect of pH and temperature on the growth of isolated strains. Here, (a) and (b) indicate the effect of pH on the growth of S-11 and S-15, respectively. (c) and (d) indicates the effect of temperature on the growth of S-11and S-15, respectively. Different letters indicate the significance differences at a p < 0.05 significance level.
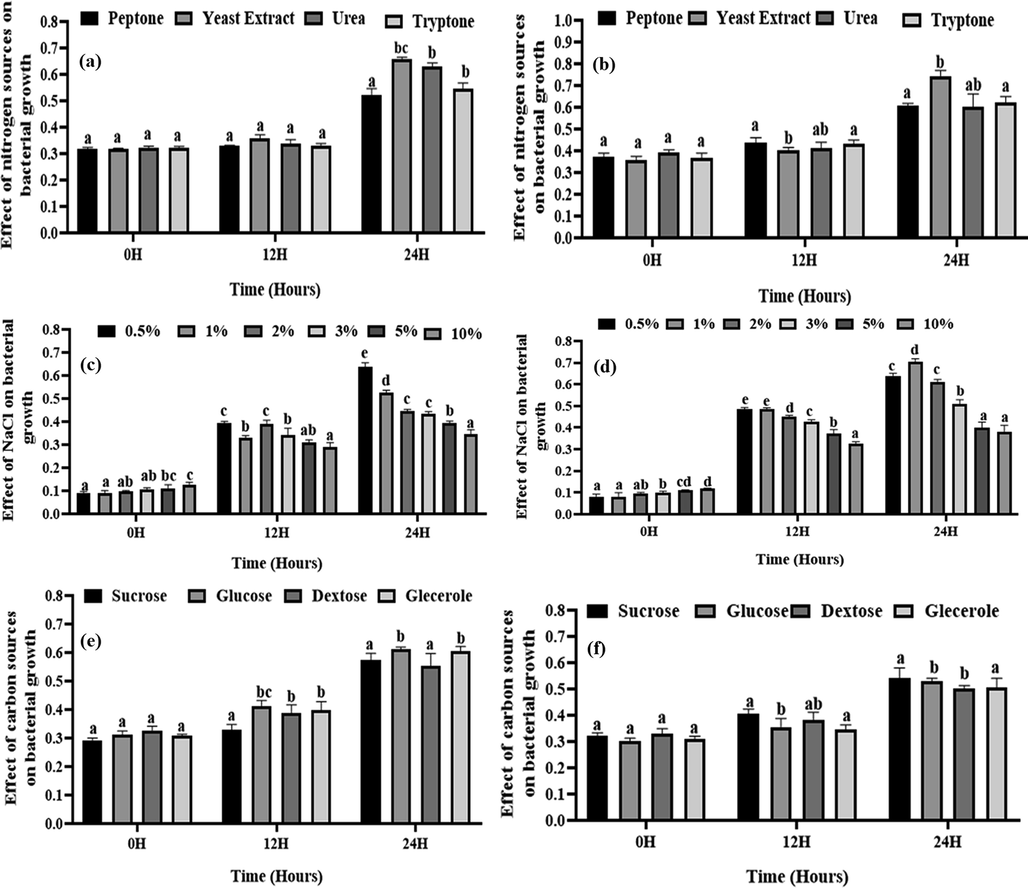
Effect of nitrogen, NaCl concentration, and carbon, on isolated bacterial growth. Here, (a) and (b) indicates the effect of nitrogen sources on growth of S-1 and S-15, respectively; (c) and (d) indicates the effect of NaCl concentrations on the growth of S-11and S-15, respectively; (e) and (f) indicates the effect of carbon source on the growth of S-11and S-15, respectively. Different letters indicate the significance differences at a p < 0.05 significance level.
3.4 Mixed dyes degradation
In the case of mixed dyes like MG + BV, the degradation rate was 74% for S-11 and 75% for S-15, with MG + AB, it was 75% for S-11 and 82% for S-15, with AB + BV, the degradation rate was 73% for S-11 and 73% for S-15, and finally, with a triple combination of MG + BV + AB, the degradation rate was 80% for S-11 and 82% for S-15 (Fig. 4 and supplementary Table-2 ). Isolate S-15 can degrade more frequently when the mixture of dyes remain MG + BV + AB and MG + AB than S-11 and others combinations of dyes.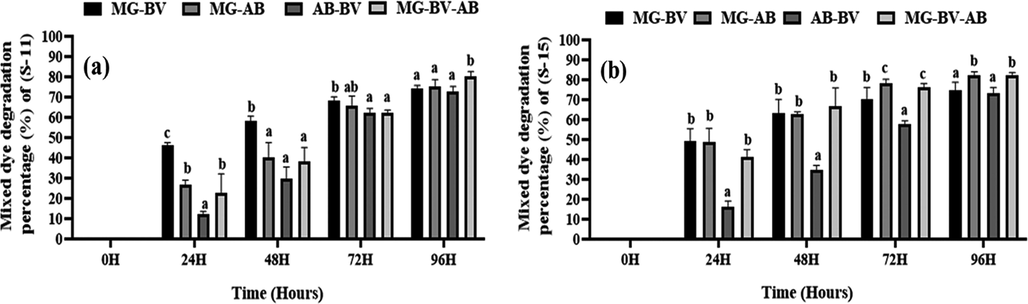
Mixed dye degradation rate of isolated bacterial strains. Here, (a) and (b) indicate the degradation rate of mixed dyes (double and triple) by S-11 and-15, respectively. M−G, A-B, and B-V indicate methylene green, acid blue, and basic violet dye, respectively. Different letters indicate the significance differences at a p < 0.05 significance level.
3.5 Morphological, biochemical, and antibiotic sensitivity test
The morphological and biochemical tests results of the isolated S-11 and S-15 are shown in supplementary Table-3 and the antibiotic sensitivity was shown in supplementary Table-4 and supplementary Fig. 2. Among twelve antibiotics S-11 showed resistance to five antibiotics and S-15 showed resistance to six antibiotics.
3.6 Molecular identification
The isolates S-11 and S-15 shared a 96.99% similarity with Bacillus pseudomycoides and a 98.28% similarity with Acinetobacter haemolyticus, respectively in molecular identification. These strains were also used to build a phylogenetic tree (Supplementary Fig 7).
3.7 Molecular docking
The docking study was conducted to explore the binding interactions of the azoreductase and three different dyes. The methylene green, basic violet, and acid blue had binding energy of −6.3, −6.6, and −6.8 Kcal/mol, respectively while interacting with the azoreductase (Table-1 and Fig. 5). The methylene blue and azoreductase protein form one alkyl bond at Pro101, and two pi-alkyl bonds at His10 and Trp103. This compound made two contacts at active sites of the Azoreductase protein; His10, and Trp103. The Basic violet and azoreductase protein were stabilized by the interactions of two hydrogen bonds at Asp116, Lys112, while four pi-alkyl bonds at Asp116, Phe57, Ala119, and Ilu169 residues and two alkyl bond at Ile52, Ile168, and two pi-pi-T shaped interactions at Trp60 (active site) and Phe172 residues. The acid blue and target protein was stabilized by the interactions of one hydrogen bond at Asn104 (active site), one pi-sulfur at His186 (active site), one alkyl bond at Pro11, and two pi-alkyl interactions at His10 and Phe18.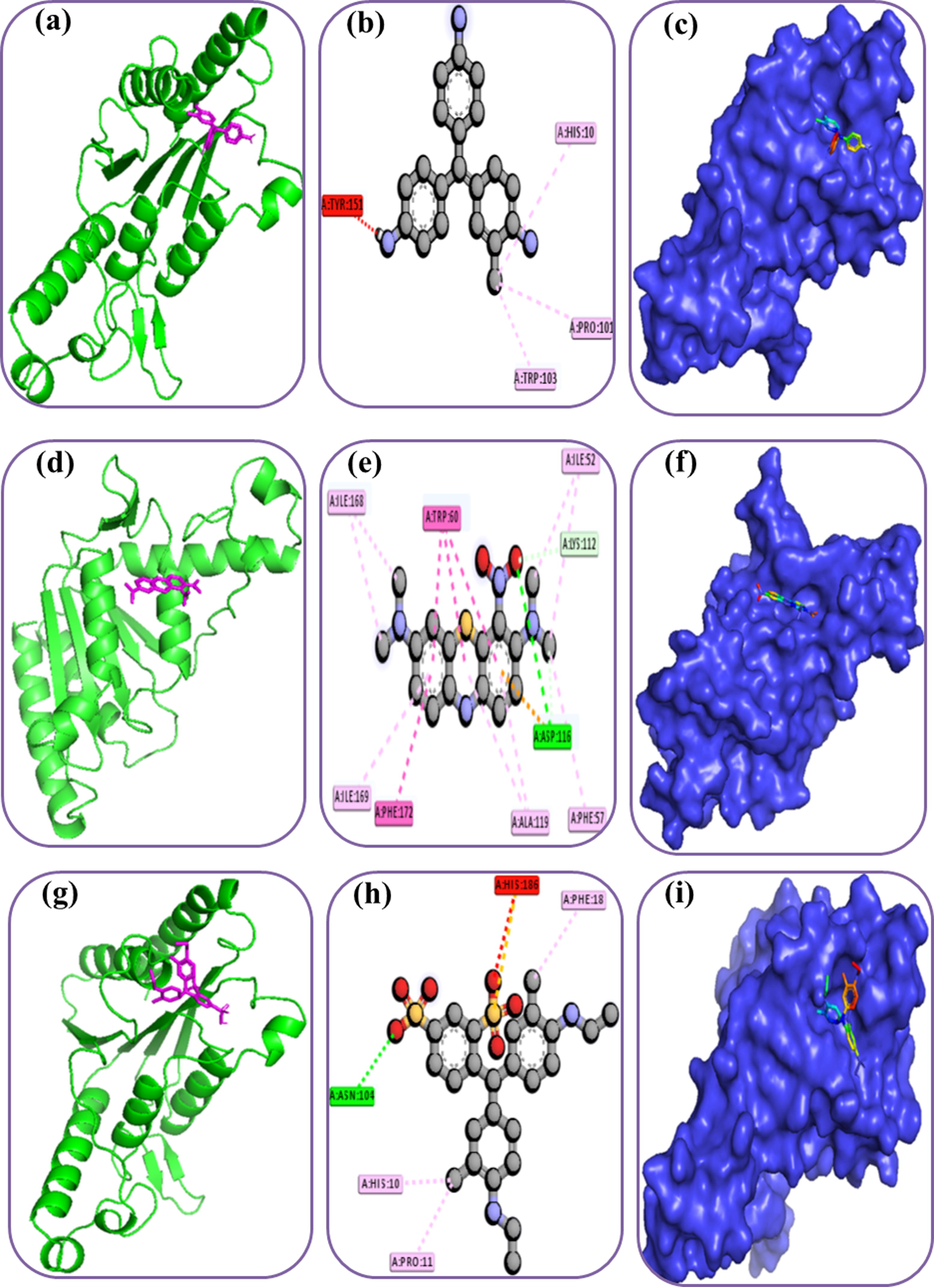
Docking simulation between Azoreductase proteins with the different dyes. Here, (a), (b), (c) indicates the Cartoon, 3D, surface view of the protein and Methylene Green dye complex; (d), (e), (f) indicates the Cartoon, 3D, surface view of the protein and basic violet dye complex; (g), (h), (i) indicates the Cartoon, 3D, surface view of the protein and acid blue dye complex.
3.8 Molecular dynamics
The molecular dynamics study was conducted to understand the stable behavior of the docked complexes. The root means square deviations of the complexes were explored to understand the flexibility of the complexes across the simulation trajectories.
Fig. 6(a) indicated that the RMSD of acid blue, methylene green, basic violet had the upper trend in the very beginning of simulation periods, which might be responsible for the flexible nature of the complexes. Therefore, the three complexes had reached the steady-state after 20 ns and maintained the lower degree of deviations, which defines the structural integrity of the complexes. Moreover, The RMSD of all three complexes was<2.5 Å, indicating that the complexes were structurally stiff.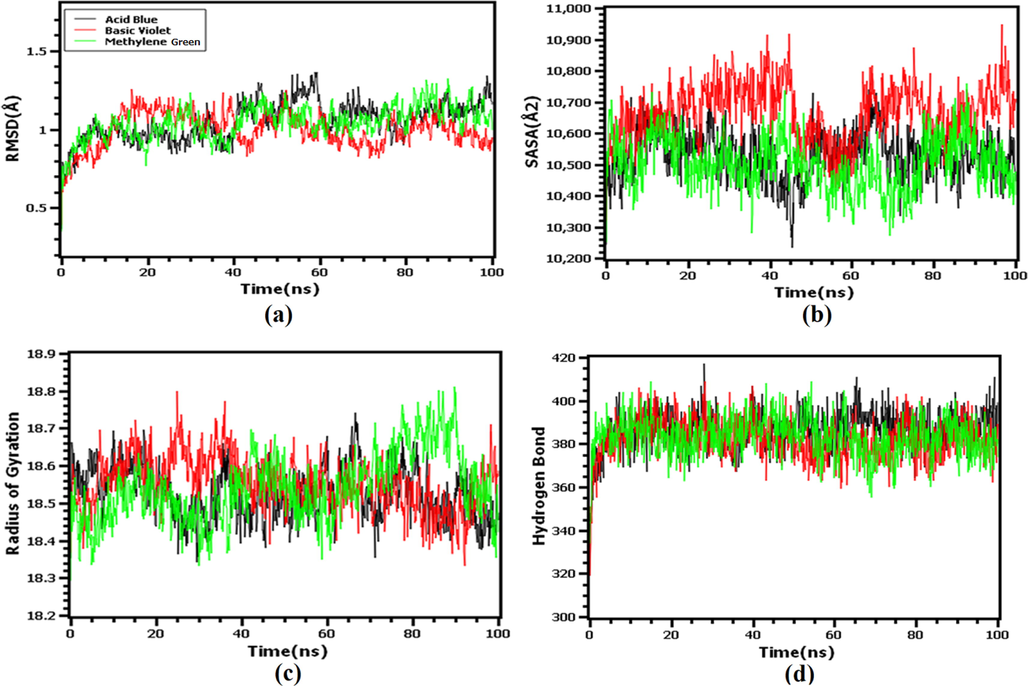
The molecular dynamics simulation of the docked complexes. Here (a) Root mean square deviation of the three docked complexes, (b) solvent accessible surface area, (c) radius of gyration, and (d) hydrogen bond of the docked complexes.
Aside from the solvent-accessible surface area of the complexes, the SASA of the complexes was also analyzed, with a higher SASA indicating a large surface area and a lower SASA indicating a truncated nature of the complexes. Fig. 6(b) indicated that the basic violet complexes had increased SASA after interaction with the protein, which corresponded to the expansion of the surface area. The other two complexes had lower and stable SASA profiles, indicating the stable nature of the complexes.
Therefore, the radius of gyrations of the systems was also explored, with higher Rg indicating a more mobile nature and lower Rg indicating a more steady nature of the complexes. According to Fig. 6(c), the complexes had a stable Rg profile throughout the simulated trajectories and did not fluctuate substantially. The hydrogen bond pattern of the three systems was stable and had a lower degree of deviations, indicating that the complexes were stable [Fig. 6(d)].
4 Discussion
Textile dyes utilize a lot of water, which results in effluents. This effluent contaminates the water surface in the vicinity of the city. When dyes are exposed to light and washed, these colors remain exceedingly stable. As a result, they maintain their color and structural integrity in the environment for a long period (Sun et al., 2020). The bright hue of discharged dyes has a tremendous impact on the aquatic ecosystem, even at extremely low concentrations. Light penetration and photosynthesis are reduced as a result, and the release of such compounds has a long-term impact on the system's ecological balance. On plants and wildlife, certain dyes and their degradation compounds are poisonous and mutagenic (Pinheiro et al., 2004). To eliminate the dye toxicity from textile effluent, industrial effluents containing azo dyes must be treated before being discharged into the environment (Kishor et al., 2021). As a result, the textiles sector relies heavily on the isolation, characterization, and degrading capabilities of different bacterial strains. According to the reports, various microorganisms can degrade dyes, but only a few strains can withstand the conditions of dyeing effluents, suggesting that effluent-adapted strains may be stronger candidates for bioremediation applications (Thangaraj et al., 2022). Current finding proves that bacteria are naturally adaptable in textile effluents and capable of degrading various kinds of dyes of textile industry wastewaters. In this study, the isolated B. pseudomycoides and A. haemolyticus demonstrated significant methylene green, acid blue, and basic violet dye degradation. Abrar Ahmed et al., (2009), reported that cultures of Bacillus odysseyi SUK3, Morganella morganii SUK5, Proteus sp. SUK7, could decolorize 50 mg/l of reactive blue 59 by 82%, 90%, and 89%, at 24, 30, and 60 h respectively. Bhattacharya et al., (2017) also reported that Nesterenkonia lacusekhoensis EMLA3 showed 83% of dye removal in alkaline and salt-rich dyeing effluent after 120 h of treatment under static conditions. The dye can be degraded successfully by single microbial culture was reported by Kapoor et al., (2021).
Single dye degradation by bacteria is a well-known study in biodegradation, where mixed dye degradation has gotten significantly less attention. In this study, significant result was observed in case of mixed dye degradation for both B. pseudomycoides and A. haemolyticus strains. Rajeswari et al., (2013) found that the five most efficient strains (TU57, TR26, VP9, KP27, and KP23) out of 112 were capable of decolourizing 2700 ppm mixed dyes during 13 days of incubation and KP23 was able to degrade 100 ppm mixed dyes within 24 h of incubation. Bacterial growth patterns differ greatly across the species (Yuan et al., 2021). In this investigation, B. pseudomycoides and A. haemolyticus were found to vary in concentrations and conditions of nutrient sources which was similar to a previous study reported by del Rio et al. (2016). Our findings are also slightly closer to the previous studies by Buthelezi et al. (2012) and Lalnunhlimi & Veenagayathri (2016).
Antibiotic-resistant bacteria and genes are abundant in wastewater treatment plants, which can be passed on to other bacteria in the environment (Osińska et al., 2020). Therefore, the detection of the antibiotic resistance profile of the chosen bacteria is pivotal for large-scale implementation of these bacteria in treatment plants. Current study showed that, B. pseudomycoides was resistant to penicillin, amoxicillin, cefuroxime, cefixime & Ceftazidime and A. haemolyticus resistant to penicillin, amoxicillin, cefuroxime, ampicillin cefixime & ceftazidime. Roy et al. (2020) isolated antibiotic-resistant Enterobacter spp. CV-S1 and CM-S1 from textile effluents, where both the strains were resistant to bacitracin, cephradine, and erythromycin (Roy et al., 2020).
Moreover, the docking study was conducted to explore the binding interactions between the bacterial azoreductase protein and the three dyes. The bacterial azoreductase protein is responsible for breaking down the azo bond fund in the textile azo dyes. Consequently, bacteria would be aided in degrading the dyes. Computational approaches could help by predicting the toxicity and nature of the interacting target receptor protein with the ligand. Molecular docking is extensively used in all sectors of applied biological sciences in optimizing the interaction and significance among protein-ligands (Sridhar and Helan Chandra, 2014).
Integrating homology modeling, molecular docking and protein-protein interaction studies can be performed to screen and elucidate the structure and function of enzymes (de Ruyck et al., 2016). Molecular docking aids bioremediation strategies by forecasting various parameters such as ligand and protein nature, interaction, theoretical mechanisms, and toxicity for an efficient technology transfer to real time set-up (Sridhar and Helan Chandra, 2014). It can be used to be predict and screen pollutants for their proclivity for the bioremediation by available enzymes (Suresh et al., 2008). The in-silico approach can predict the chemical nature of a contaminant, novel xenobiotic biodegradation pathways, and microorganisms capabilities of biotransformation at the environmental levels (Sarkar et al., 2012).
Finally, Kumar et al. (2019) reported that B. pseudomycoides was able to degrade azo dye acid black 24 promisingly in a study of decolorization. In another study found that A. haemolyticus degraded textile industrial dyes using a mixture of gold nanocatalysts (Wadhwani et al., 2018). Both the studies together with most of the previously cited studies concentrated on single dye degradation only and focus on mixed dyes degradation was neglected.
5 Strengths and limitations of this study
Most of the previous research showed only single dye degradation, but our study focuses on both single and mixed dye degradation capability with in silico binding interaction and stability. We have identified two bacterial strains which are able to degrade single dye as well as mix dyes with high efficacy. Furthermore, molecular docking and simulation was conducted to explore the binding interactions and stability between the bacterial azo reductase protein and the three dyes which provide significance values in this research study. Determination of the end products of degraded dye compounds and the bacterial gene/genes responsible for degradation were not considered, that could be the limitation of current study.
6 Conclusion
Bioremediation is now a promising technology for removing contaminants from the environment that is eco-friendly, cost-effective, and simple to implement that help to improve the quality of the water ecosystem. In this research, all eight isolated bacterial strains were able to degrade MG, AB, and BV dyes where Bacillus pseudomycoides, and Acinetobacter haemolyticus exhibited remarkable single and mixed dye decolorization capacity. As a result, these two strains can be employed as a bioremediation agent in the treatment of textile dye polluted wastewater and soils to achieve biodegradation and reduce the toxicity of textile dyes. The ability of these bacteria can be used to extract residual dyes from the sources of wastewater for environmental cleanup and ecosystem restoration. Many textiles industry discharge their waste dye effluents into the environment which remain in the environment for longer period and do not degrade because of their stuck aromatic structure. So, presence of azo dye in the environment creates several difficulties that endanger aquatic and terrestrial life. These effluents are usually treated using through physical, chemical, and biological methods before being discharged. The employment of these methods to treat dye effluents is insufficient since, in most cases, partial degradation occurs, resulting in the production of many hazardous metabolites and also costly and not eco-friendly. The findings of molecular docking and dynamic simulations were also satisfactory, predicting the molecular level mechanism. The toxicity of degraded products is being investigated. However, further research is needed to detect the end products of the dye compounds as well as discover the genes responsible for the decolorization of textile azo dyes.
Acknowledgements
Ministry of Science and Technology, Bangladesh
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Isolation, identification, and characterization of cadmium resistant pseudomonas sp. M3 from industrial wastewater. J. Waste Manag.. 2014;2014:1-6.
- [CrossRef] [Google Scholar]
- Biological efficiency and nutritional contents of Pleurotus florida (Mont.) singer cultivated on different agro-wastes. Nat. Sci.. 2009;7:44-48.
- [Google Scholar]
- Scenario analysis of textile industry in asia-pacific trade agreement (APTA) Procedia Comput. Sci.. 2017;122:685-690.
- [CrossRef] [Google Scholar]
- Optimization of triazo Acid Black 210 dye degradation by Providencia sp. SRS82 and elucidation of degradation pathway. Process Biochem.. 2014;49:110-119.
- [CrossRef] [Google Scholar]
- The protein data bank. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002
- [CrossRef] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E., 2000. The Protein Data Bank 28, 235–242
- Emerging and eco-friendly approaches for waste management. Emerg. Eco-Friendly Approaches Waste Manag.. 2018;1–435
- [CrossRef] [Google Scholar]
- Degradation of azo dye methyl red by alkaliphilic, halotolerant Nesterenkonia lacusekhoensis EMLA3: application in alkaline and salt-rich dyeing effluent treatment. Extremophiles. 2017;21:479-490.
- [CrossRef] [Google Scholar]
- BIOVIA, D., 2016. Discovery Studio Modeling Environment, Release 2017, San Diego: DassaultSystèmes, 2016. Adres http//accelrys. com/products/collaborative-science/biovia-discoverystudio/visualization download. php.
- Textile dye removal from wastewater effluents using bioflocculants produced by indigenous bacterial isolates. Molecules. 2012;17:14260-14274.
- [CrossRef] [Google Scholar]
- Case, D.A., Babin, V., Berryman, J.T., Betz, R.M., Cai, Q., Cerutti, D.S., Cheatham III, T.E., Darden, T.A., Duke, R.E., Gohlke, H., Goetz, A.W., Gusarov, S., Homeyer, N., Janowski, P., Kaus, J., Kolossváry, I., Kovalenko, A., Lee, T.S., LeGrand, S., Luchko, T., Luo, R., Madej, B., Merz, K.M., Paesani, F., Roe, D.R., Roitberg, A., Sagui, C., Salomen-Ferrer, R., Seabra, G., Simmerling, G.L., Smith, W., Swails, J., Walker, R.C., Wang, J., Wolf, R.M., Wu, X., Kollman, P.A., 2014. AMBER14. AMBER 14.
- Waste management by biological approach employing natural substrates and microbial agents for the remediation of dyes’ wastewater. Appl. Sci.. 2020;10
- [CrossRef] [Google Scholar]
- Molecular docking as a popular tool in drug design, an in silico travel. Adv. Appl. Bioinforma. Chem.. 2016;9
- [CrossRef] [Google Scholar]
- Putrescine production by Lactococcus lactis subsp. cremoris CECT 8666 is reduced by NaCl via a decrease in bacterial growth and the repression of the genes involved in putrescine production. Int. J. Food Microbiol.. 2016;232:1-6.
- [CrossRef] [Google Scholar]
- Microplastic pollution in water and sediment in a textile industrial area. Environ. Pollut.. 2020;258:113658
- [CrossRef] [Google Scholar]
- Review paper on current technologies for decolourisation of textile wastewaters: perspectives for anaerobic biotechnology. Bioresour. Technol.. 2007;98:2369-2385.
- [CrossRef] [Google Scholar]
- Phytochemicals from leucas zeylanica targeting main protease of sars-cov-2: chemical profiles, molecular docking, and molecular dynamics simulations. Biology (Basel). 2021;10
- [CrossRef] [Google Scholar]
- A smooth particle mesh Ewald method. J. Chem. Phys.. 1995;103:8577-8593.
- [CrossRef] [Google Scholar]
- The Potential of ZrO2 catalyst toward degradation of dyes and phenolic compound. Mater. Today Proc.. 2019;19:1524-1528.
- [CrossRef] [Google Scholar]
- Fletcher, C.A., St. Clair, R., Sharmina, M., 2021. A framework for assessing the circularity and technological maturity of plastic waste management strategies in hospitals. J. Clean. Prod. 306, 127169. Doi: 10.1016/j.jclepro.2021.127169.
- Gürses, A., Açıkyıldız, M., Güneş, K., Gürses, M.S., 2016. Colorants in Health and Environmental Aspects 69–83. Doi: 10.1007/978-3-319-33892-7_5.
- Merck molecular force field. I. basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem.. 1996;17:490-519.
- [CrossRef] [Google Scholar]
- Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. J. Comput. Chem.. 1996;17:520-552.
- [CrossRef] [Google Scholar]
- Structure and dynamics of TIP3P, TIP4P, and TIP5P water near smooth and atomistic walls of different hydroaffinity. J. Chem. Phys.. 2014;140:174501
- [Google Scholar]
- An implementation of the smooth particle mesh Ewald method on GPU hardware. J. Chem. Theory Comput.. 2009;5:2371-2377.
- [CrossRef] [Google Scholar]
- Textile dyeing industries in Bangladesh for sustainable development. Int. J. Environ. Sci. Dev.. 2011;428–436
- [CrossRef] [Google Scholar]
- 1001 Ways to run AutoDock Vina for virtual screening. J. Comput. Aided. Mol. Des.. 2016;30:237-249.
- [CrossRef] [Google Scholar]
- Subcritical and supercritical water oxidation for dye decomposition. J. Environ. Manage.. 2021;290:112605
- [CrossRef] [Google Scholar]
- A comparison of extended spectrum β-lactamase producing Escherichia coli from clinical, recreational water and wastewater samples associated in time and location. PLoS One. 2017;12:1-15.
- [CrossRef] [Google Scholar]
- Synergistic role of bacterial consortium to biodegrade toxic dyes containing wastewater and its simultaneous reuse as an added value. Chemosphere. 2021;284:131273
- [CrossRef] [Google Scholar]
- Industrial effluents and their impact on water quality of receiving rivers in Nigeria. J. Applieal Technol. Environ. Sanit.. 2011;1:75-86.
- [Google Scholar]
- Exploiting microbial biomass in treating azo dyes contaminated wastewater: mechanism of degradation and factors affecting microbial efficiency. J. Water Process Eng.. 2021;43
- [CrossRef] [Google Scholar]
- Ecotoxicological and health concerns of persistent coloring pollutants of textile industry wastewater and treatment approaches for environmental safety. J. Environ. Chem. Eng.. 2021;9:105012
- [CrossRef] [Google Scholar]
- Assignment of protonation states in proteins and ligands: Combining pK a prediction with hydrogen bonding network optimization. Methods Mol. Biol.. 2012;819:405-421.
- [CrossRef] [Google Scholar]
- Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model.. 2006;25:481-486.
- [CrossRef] [Google Scholar]
- New ways to boost molecular dynamics simulations. J. Comput. Chem.. 2015;36:996-1007.
- [CrossRef] [Google Scholar]
- Decolorization of azo dyes (Direct Blue 151 and Direct Red 31) by moderately alkaliphilic bacterial consortium. Brazilian J. Microbiol.. 2016;47:39-46.
- [CrossRef] [Google Scholar]
- YASARA: A tool to obtain structural guidance in biocatalytic investigations. Methods Mol. Biol.. 2018;1685:43-67.
- [CrossRef] [Google Scholar]
- Tartrazine: physical, thermal and biophysical properties of the most widely employed synthetic yellow food-colouring azo dye. J. Therm. Anal. Calorim.. 2018;134:209-231.
- [CrossRef] [Google Scholar]
- Microalgae-based wastewater treatment for nutrients recovery: a review. Bioresour. Technol.. 2019;291:121934
- [CrossRef] [Google Scholar]
- Sulfonates: novel electron accepters in anaerobic respiration. Arch. Microbiol.. 1996;166:204-210.
- [CrossRef] [Google Scholar]
- Dye-decolorization of a newly isolated strain Bacillus amyloliquefaciens W36. World J. Microbiol. Biotechnol.. 2021;37
- [CrossRef] [Google Scholar]
- Decolorization of synthetic dyes using bacteria isolated from textile industry effluent. Asian J. Biotechnol.. 2012;4:129-136.
- [CrossRef] [Google Scholar]
- Mahmud, S., Biswas, S., Kumar Paul, G., Mita, M.A., Afrose, S., Robiul Hasan, M., Sharmin Sultana Shimu, M., Uddin, M.A.R., Salah Uddin, M., Zaman, S., Kaderi Kibria, K.M., Arif Khan, M., Bin Emran, T., Abu Saleh, M., 2021a. Antiviral peptides against the main protease of SARS-CoV-2: A molecular docking and dynamics study. Arab. J. Chem. 14, 103315. Doi: 10.1016/j.arabjc.2021.103315.
- Mahmud, S., Biswas, S., Paul, G.K., Mita, M.A., Promi, M.M., Afrose, S., Hasan, R., Zaman, S., Uddin, S., Dhama, K., 2021b. Plant-Based Phytochemical Screening by Targeting Main Protease of SARS-CoV-2 to Design Effective Potent Inhibitors 1–18.
- Prospective role of peptide-based antiviral therapy against the main protease of SARS-CoV-2. Front. Mol. Biosci.. 2021;8
- [CrossRef] [Google Scholar]
- Manzoor, J., Sharma, M., 2019. Impact of Textile Dyes on Human Health and Environment 162–169. Doi: 10.4018/978-1-7998-0311-9.ch008.
- Mishra, S., Nayak, J.K., Maiti, A., 2020. Bacteria-mediated bio-degradation of reactive azo dyes coupled with bio-energy generation from model wastewater. Clean Technol. Environ. Policy 22, 651–667. Doi: 10.1007/s10098-020-01809-y.
- Fed-batch decolorization of poly R-478 by Trametes versicolor. Brazilian Arch. Biol. Technol.. 2004;47:179-183.
- [CrossRef] [Google Scholar]
- Mousavi, S.S., Karami, A., Haghighi, T.M., Tumilaar, S.G., Fatimawali, Idroes, R., Mahmud, S., Celik, I., Ağagündüz, D., Tallei, T.E., Emran, T. Bin, Capasso, R., 2021. In silico evaluation of iranian medicinal plant phytoconstituents as inhibitors against main protease and the receptor-binding domain of sars-cov-2. Molecules 26. Doi: 10.3390/molecules26185724.
- Ogola, H.J.O., Ashida, H., Ishikawa, T., Sawa, Y., 2015. Explorations and Applications of Enzyme-linked Bioremediation of Synthetic Dyes. Adv. Bioremediation Wastewater Polluted Soil. Doi: 10.5772/60753.
- Small-scale wastewater treatment plants as a source of the dissemination of antibiotic resistance genes in the aquatic environment. J. Hazard. Mater.. 2020;381
- [CrossRef] [Google Scholar]
- Isolation and characterization of bacteria from two soil samples and their effect on wheat (Triticum aestivum l.) growth promotion. J. Adv. Biotechnol. Exp. Ther.. 2020;3:254-262.
- [CrossRef] [Google Scholar]
- Aromatic amines from azo dye reduction: Status review with emphasis on direct UV spectrophotometric detection in textile industry wastewaters. Dye Pigment. 2004;61:121-139.
- [CrossRef] [Google Scholar]
- Fermentation optimization of cellulase production from sugarcane bagasse by Bacillus pseudomycoides and molecular modeling study of cellulase. Curr. Res. Microb. Sci.. 2021;2
- [CrossRef] [Google Scholar]
- Lithium ion conducting solid polymer blend electrolyte based on bio-degradable polymers. Bull. Mater. Sci.. 2013;36:333-339.
- [CrossRef] [Google Scholar]
- Roy, D.C., Sheam, M., Hasan, R., Saha, A.K., Roy, A.K., Haque, E., Rahman, M., Swee-Seong, T., Kumar Biswas, S., 2020. Isolation and characterization of two bacterial strains from textile effluents having Malachite Green dye degradation ability. bioRxiv 1–15.
- In-silico feasibility of novel biodegradation pathways for1-naphthyl methylcarbamate. J. Toxicol. Sci.. 2012;4:89-93.
- [Google Scholar]
- Liquid serial dilution is inferior to solid media for isolation of cultures representative of the phylum-level diversity of soil bacteria. Appl. Environ. Microbiol.. 2004;70:4363-4366.
- [CrossRef] [Google Scholar]
- Involvement of computational tools towards in silico remediation - synthetic textile dyes interacting with azoreductase. Int. J. ChemTech Res.. 2014;6:4412-4416.
- [Google Scholar]
- In silico analysis of bacterial systems for textile azo dye decolorization and affirmation with wetlab studies. Clean - Soil, Air, Water. 2017;45
- [CrossRef] [Google Scholar]
- Lightweight graphene oxide-based sponges with high compressibility and durability for dye adsorption. Carbon N. Y.. 2020;160:54-63.
- [CrossRef] [Google Scholar]
- An Insilco approach to bioremediation: laccase as a case study. J. Mol. Graph. Model.. 2008;26:845-849.
- [CrossRef] [Google Scholar]
- Heavy metal determination and aquatic toxicity evaluation of textile dyes and effluents using Artemia salina. Biocatal. Agric. Biotechnol.. 2020;25
- [CrossRef] [Google Scholar]
- Biodegradation of Reactive Red 198 by textile effluent adapted microbial strains. Arch. Microbiol. 2022;204(1):1-13.
- [Google Scholar]
- AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009
- [CrossRef] [Google Scholar]
- the effect of growth parameters on the antibiotic activity and sporulation in Bacillus Spp. isolated from soil. J. Microbiol. Biotechnol. Food Sci.. 2018;8:2310-2313.
- [Google Scholar]
- Eradication of fatal textile industrial dyes by wastewater treatment. Biointerface Res. Appl. Chem.. 2021;12:567-587.
- [CrossRef] [Google Scholar]
- Comparing AutoDock and Vina in ligand/decoy discrimination for virtual screening. Appl. Sci. 2019
- [CrossRef] [Google Scholar]
- Wadhwani, S.A., Shedbalkar, U.U., Nadhe, S., Singh, R., Chopade, B.A., 2018. Decolorization of textile dyes by combination of gold nanocatalysts obtained from Acinetobacter sp. SW30 and NaBH4, Environmental Technology and Innovation. Doi: 10.1016/j.eti.2017.12.001.
- Bioreduction (AuIII to Au0) and stabilization of gold nanocatalyst using Kappa carrageenan for degradation of azo dyes. Int. J. Biol. Macromol. 2021;176:282-290.
- [Google Scholar]
- Development and testing of a general Amber force field. J. Comput. Chem.. 2004;25:1157-1174.
- [CrossRef] [Google Scholar]
- Fungal-bacterial cooccurrence patterns differ between arbuscular mycorrhizal fungi and nonmycorrhizal fungi across soil niches. MBio. 2021;12
- [CrossRef] [Google Scholar]
- Feasibility and applicability of the scaling-up of bio-electro-Fenton system for textile wastewater treatment. Environ. Int.. 2020;134:105352
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2022.104078.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary data 1
Supplementary data 1
Supplementary data 2
Supplementary data 2







