Translate this page into:
Network pharmacological analysis of ethanol extract of Morus alba linne in the treatment of type 2 diabetes mellitus
⁎Corresponding authors. changshui@hotmail.com (Xiao Zhang), leonwang@xzhmu.edu.cn (Liang Wang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Diabetes is a chronic endocrine metabolism disorder that leads to hyperglycaemia. As the most common diabetic type, type 2 diabetes mellitus accounts for 90% of all diabetic disease with a key feature of insulin resistance. Mulberry leaves are commonly used in traditional Chinese medicine, and many studies confirm that mulberry leaves have positive effects on alleviating the pathological conditions of type 2 diabetes. However, its anti-diabetic effects and active ingredients are not completely understood. Compared with time- and cost-consuming experiments, network pharmacology provided a convenient method to systematically investigate the interactions between compounds in mulberry leaves and diabetes-related genes. Although similar network pharmacological studies were performed for mulberry leaves, only compounds sourced from public database or volatile components in the mulberry leaves were examined. In this study, we initially analysed the bioactive compounds from ethanol extract of mulberry leaves via liquid chromatography coupled with mass spectrometry, which generated a total of 248 components. A total of 14 active components and 49 potentially functional compounds were identified from TCMSP and PharmMapper database, respectively, which worked on 37 target proteins directly involved in the pathogenesis of T2DM. Kaempferol was shown to be the most influential active compound while androgen receptor was most widely regulated by compounds in the ethanol extract of mulberry leaves. Moreover, protein–protein interaction and target-pathway network analysis revealed that AKT1 was most important in the 37 target proteins in terms of its functions in different pathways and interactions with other target proteins. Moreover, pathway analysis showed that ethanol extract of mulberry leaves alleviated type 2 diabetes mellitus through pathways such as TNF signaling pathway, NF-κB signaling pathway, and insulin resistance pathway. In sum, this study provided a complete overview of the working mechanisms for the ethanol extract of mulberry leaves, the results of which could be used as a practical guidance for further experimental investigation of the functions of mulberry leaves during the treatment of type 2 diabetes mellitus.
Keywords
Mulberry leaves
Network pharmacology
Diabetes
LC-MS
Ethanol extraction
- AEML
-
Aqueous Extracts of Mulberry Leaf
- AKT1
-
RAC-alpha serine/threonine-protein kinase
- BCL2
-
Apoptosis regulator Bcl-2
- DAVID
-
Database for Annotation, Visualization and Integrated Discovery
- DL
-
Drug Likeness
- DM
-
Diabetes Mellitus
- EEML
-
Ethanol Extract of Mulberry Leaves
- GO
-
Gene Ontology
- HPLC
-
High Performance Liquid Chromatography
- IKBKB
-
Kappa-B Kinase Subunit Beta
- IR
-
Insulin Resistance
- KEGG
-
Kyoto Encyclopedia of Genes and Genomes
- KOBAS
-
KEGG Orthology Based Annotation System
- LC-MS
-
Liquid Chromatography Coupled with Mass Spectrometry
- Mcode
-
Molecular Complex Detection
- MLE
-
Mulberry Leaf Extract
- MS
-
Mass Spectrometry
- OB
-
Oral Bioavailability
- OMIM
-
Online Mendelian Inheritance in Man
- PPI
-
Protein-Protein Interaction
- SMBG
-
Self-monitoring Blood Glucoses
- T1DM
-
Type 1 Diabetes Mellitus
- T2DM
-
Type 2 Diabetes Mellitus
- TCM
-
Traditional Chinese Medicine
- TCMSP
-
Traditional Chinese Medicine Systems Pharmacology
- TLR-2
-
Toll-like Receptor 2
- TNF-α
-
Tumour Necrosis Factor-α
- UPLC-MS/MS
-
Ultra-Performance Liquid Chromatography-Tandem Mass Spectrometry
Abbreviations
1 Background
Diabetes mellitus (DM) is a complex metabolic disorder, which is mainly divided into two types, that is, Type 1 Diabetes Mellitus (T1DM) caused by insulin secretion defects and Type 2 Diabetes Mellitus (T2DM) due to insulin resistance. One of the common features of diabetes is the long-term chronic hyperglycaemia that will finally develop into end organ failures (Tsalamandris et al., 2019). In China, diabetes was first recorded in an ancient Chinese medical book The Yellow Emperor’s Classic of Internal Medicine (Yu et al., 2018). Since then, traditional Chinese medicine (TCM) has been used to treat the disease for over 2000 years. As an integral part of TCM, herbal medicines are widely used in the treatment of diabetes, such as Radix rehmanniae and Momordica charantia, etc. (Wang et al., 2013). Herbs function differently when used to alleviate the conditions of diabetes, some of which decrease blood glucose level through enhancing carbohydrate utilization while others aim to reduce inflammation and increase insulin sensitivity via certain signaling pathways (Yu et al., 2018;Tian et al., 2019).
Mulberry leaf (Morus alba L., abbr. ML) is an important TCM herb and has been used to treat a variety of diseases since ancient China (Tian et al., 2019). Previous studies have revealed that ingestion of ML is effective in the treatment of T2DM. For example, a 3-month clinical trial conducted by Riche et al. showed that mulberry leaf extract (MLE) could significantly decrease post-prandial self-monitoring blood glucoses (SMBG) between MLE group and baseline group (Riche et al., 2017). Tian et al. studied the molecular mechanisms of MLs in T2DM therapy and found out that aqueous extracts of mulberry leaf (AEML) mitigates high blood glucose, insulin resistance (IR), and inflammation through the interactions among Toll-like Receptor 2 (TLR-2) signaling pathway, insulin signaling pathway, and tumour necrosis factor-α (TNF-α) (Tian et al., 2019). However, due to the sophisticated composition of compounds in MLs, it is rather difficult to elucidate all the mechanisms for ML effects on diabetes.
In recent years, network pharmacology has been gaining more and more attentions due to its convenience in the construction of compounds-proteins (genes)-diseases relationships and its effectiveness in understanding the regulation principles of small molecules in a high-throughput manner (Zhang et al., 2019). Thus, the new approach has the potential to transform TCM from an experience-based to evidence-based medicine through a better understanding of the interactions among biological systems, drugs, and diseases. Although a couple of previous studies explored the effects of MLs on diabetes via network pharmacology, it is either using mulberry data from public database or focusing on volatile components of MLs (Ge et al., 2018; Wu and Hu, 2020). In this study, we analysed the components of the ethanol extract of mulberry leaves (EEML) via liquid chromatography coupled with mass spectrometry (LC-MS), which identified a total of 248 compounds. Compound-modulated targets and relevant diseases were then sourced from public databases such as TCMSP, PharmMapper, GeneCards, and Online Mendelian Inheritance in Man (OMIM), etc. (Safran et al., 2010; Ru et al., 2014). A complete network in terms of EEML treatment of T2DM was constructed, together with the function and pathway enrichment analyses. Based on the network pharmacology approach, this study elucidated the specific targets and molecular signaling pathways that EEML modulated in terms of T2DM. A schematic illustration of the methodologies used in this study is present in Fig. 1.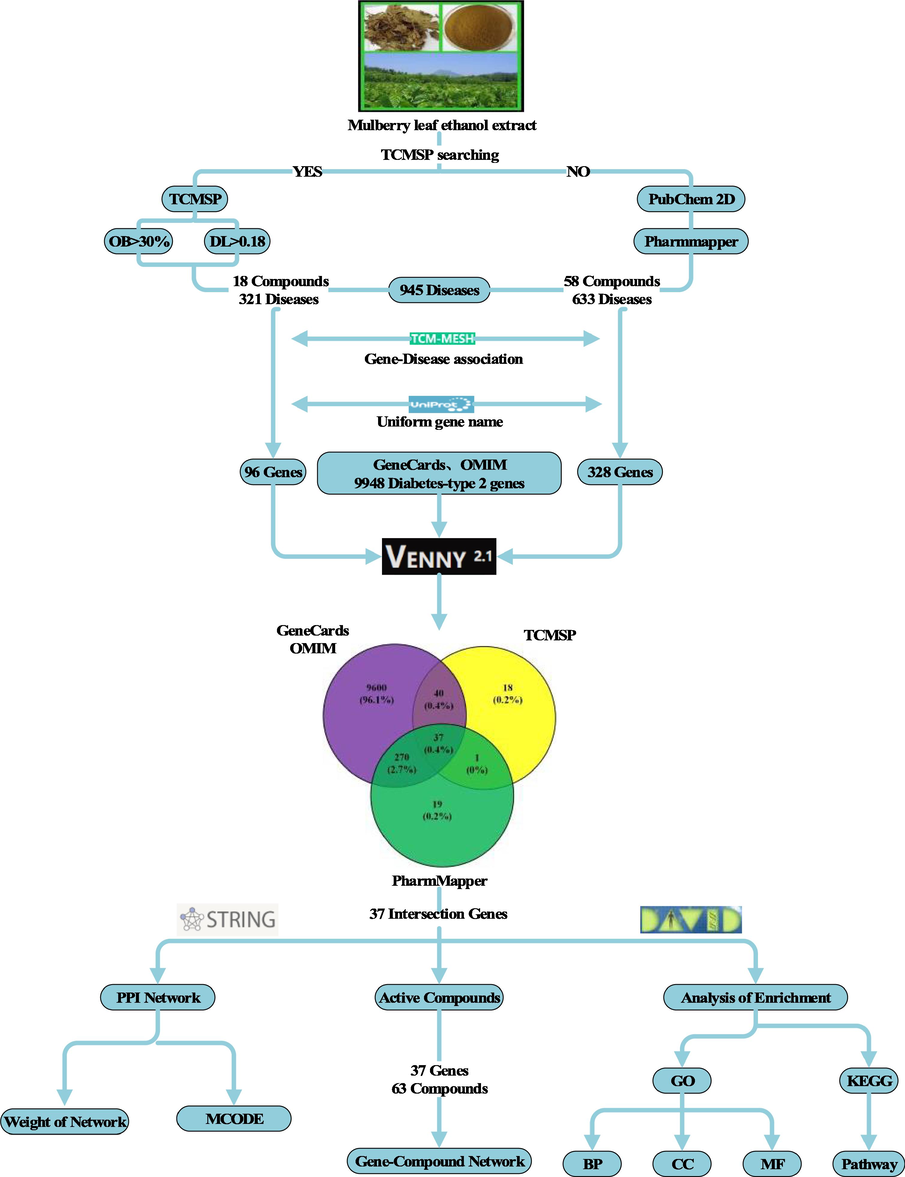
Schematic illustration of all the procedures based on network pharmacology. Based on the results of LC-MS, a set of computational tools were used for further analyses of important components in the ethanol extract of mulberry leaves and their target proteins.
2 Methods and materials
2.1 Compositions of mulberry leaves
Mulberry leaves were sourced from a local traditional Chinese medicine market in Bo-Zhou, Anhui Province, China. The plant was identified by Prof. Dao-Quan Tang from the School of Pharmacy at Xuzhou Medical University and the specimen was deposited in the Laboratory of Pharmaceutical Analysis at Xuzhou Medical University. 2 kg well-dried mulberry leaves were extracted twice with 10 kg of 70% ethanol for 90 min, which was then concentrated at 60 ℃ under vacuum condition (-0.05Mpa) to generate a total of 135 g powder through spray-drying. The powder of mulberry leaves was detected by Agilent 1100 high-performance liquid chromatography (HPLC) system (Agilent Technologies Inc., California, USA), together with Waters Xevo G2XS QT of mass spectrometry (MS) system (Waters Corporation, Milford, MA, USA). A Waters BEH C18 column (2.1 mm × 50 mm, 1.7um) was used for the separation. The mobile phase consisted of 0.1% formic acid in water (solvent A) and acetonitrile (solvent B). The gradient program started with 95% A-5% B:0–1 min, followed by 0% A-100% B:1–8 min, 0% A-100% B:8–11 min, 95% A-5% B:11–12 min. The column temperature was at 40 °C. The sample injection volume was 2 μL and the flows rate was set at 0.3 mL/min. The analytes were determined in a positive ion mode. Mass spectrometer electrospray capillary voltage was 3.0 kV, sample cone 40 V, source temperature 100 °C, desolation temperature 400 °C, cone gas 50 L/h, and desolation gas 800 L/h. A total of 248 chemical compounds were identified. A complete form of all the compounds was available in Supplementary Table S1. The corresponding total ion chromatogram was present in Supplementary Figure S1.
2.2 Compound target of EEML
We first searched traditional Chinese medicine systems pharmacology database (TCMSP, http://tcmspw.com/tcmsp.php) by using chemical name, InChIKey, and CAS Number of all the compounds identified in the ethanol extract of mulberry leaves via LC-MS. Oral bioavailability (OB, > 30%) and drug likeness (DL, > 0.18) were used as selection criteria for active ingredients and the corresponding targets of each active compound were recorded. A total of 18 compounds, together with 96 target genes were collected. Due to the limitations of the TCMSP database, we then searched the PubChem database by using chemical names in order to obtain the two-dimensional structure of each compound, which were further searched in the online database PharmMapper http://lilab-ecust.cn/Pharmmapper/ in terms of diabetes-related targets. Top 30 predicted targets for each compound were kept for further analysis. In sum, a total of 58 compounds and 327 target genes were collected.
2.3 Targets of T2DM
Target genes related to T2DM were collected through searching GeneCards and OMIM database by using the keyword Type 2 Diabetes Mellitus. 9952 candidate genes were identified. All the gene names were matched to corresponding UniProt IDs, gene names, and protein names by using ID mapping tool in the protein database UniProt (http://www.uniprot.org/). In order to construct the network between genes related with T2DM and the compounds in the ethanol extract of mulberry leaves, we pooled all the target genes and used the online tool venny (https://bioinfogp.cnb.csic.es/tools/venny/) to generate the Venn diagram (Supplementary Figure S2), which revealed that a total of 37 targets were directly associated with T2DM and targeted by 63 compounds in the ethanol extract of mulberry leaves (Table 1). Among the 63 compounds, 14 were identified from TCMSP with confirmed functions in T2DM (Table 2) while other 49 were sourced from sourced from PharmMapper (Supplementary Table S2). According to the number of interactions for the 14 components, we identified the top 5 compounds that had the highest number of interactive targets (denoted as n in parentheses for each compound) that were responsible for T2DM, which were Kaempferol (n = 23), Licoricone (n = 7), Herbacetin (n = 6), Morin (n = 5), and Lobelanidine (n = 4). Note: 1UniProt ID, gene name, and protein name were sourced from the online database https://www.uniprot.org/. 2Number of interactive components identified in the ethanol extract of mulberry leaves. 3Active components were obtained through searching TCMSP and PharmMapper databases. Note: 1Mol ID: Molecule ID used in the TCMSP database https://tcmspw.com/browse.php?qc=herbs. 2PubChem CID: Chemical compound ID used in the PubChem database https://pubchem.ncbi.nlm.nih.gov/. 3No.: Number of interactions with targets. 4Target: names of target proteins. 5OB: Oral bioavailability. 6BBB: Blood-brain barrier. 7DL: Drug-likeness. 8Structure: all the compound structures were sourced from PubChem database.
UniProt ID1
Gene Name1
Protein Name1
No.2
Interactions with Active Components3
P22303
ACHE
Acetylcholinesterase
9
13-Hydroxy-9,11-hexadecadienoic acid, 2′-Hydroxy-4,4′,6′-trimethoxydihydrochalcone, 2-Hydroxy-5-methoxy acetophenone, 6-Gingerol, Dibutyl sebacate, Heptadecylamine, Herbacetin, Kaempferol, Lupinifolin
P08588
ADRB1
Beta-1 adrenergic receptor
3
6-Gingerol, Morusimic acid B, Salsoline
P07550
ADRB2
Beta-2 adrenergic receptor
8
1,7-Bis(4-hydroxyphenyl)-hepta-4E,6E-dien-3-one, 6-Gingerol, Bis(2-ethylhexyl) phthalate, Lobelanidine, Morusimic acid B, N-cis-Feruloyl typamine, Salsoline, Zederone
P31749
AKT1
RAC-alpha serine/threonine-protein kinase
5
13-Hydroxy-9,11-hexadecadienoic acid, 4,7-Didehydroneophysalin B, Ganoderenic acid B, Isosamarcandin, Kaempferol
P09917
ALOX5
Polyunsaturated fatty acid 5-lipoxygenase
9
13-Hydroxy-9,11-hexadecadienoic acid,2′-Hydroxy-4,4′,6′-trimethoxydihydrochalcone,3-(2′-Carboxyphenyl)-4(3H)-quinazolinone, Ciwujiatone, epi-Kansenone, Ganoderenic acid B, Kaempferol, Morin, Paristerone
P10275
AR
Androgen receptor
9
12S-Hydroxyandrographolide,3,7-Dimethyloctane-1,3,6-triol,4,7-Didehydroneophysalin B, Bigelovin, Campesterol acetate, Coronaric acid, ent-Kauran-16α,17-diol, epi-Kansenone, Herbacetin
P10415
BCL2
Apoptosis regulator Bcl-2
5
13-Hydroxy-9,11-hexadecadienoic acid, 6-Gingerol, Catenarin, Kaempferol, Methyl lucidenate P
P00918
CA2
Carbonic anhydrase 2
6
13-Hydroxy-9,11-hexadecadienoic acid, 6-Gingerol, Catenarin, Kaempferol, Methyl lucidenate P
P20309
CHRM3
Muscarinic acetylcholine receptor M3
5
Bis(2-ethylhexyl) phthalate, Jangomolide, Lobelanidine, Tenuifoliside D, Zederone
Q16678
CYP1B1
Cytochrome P450 1B
6
1-Methoxy-3,7-dimethyl-2,6-octadiene,2′-Hydroxy-4,4′,6′-trimethoxydihydrochalcone, Catenarin, Kaempferol, Maglifloenone (Denudatone), Ophiopogonanone B
P00742
F10
Coagulation factor X
6
4,7-Didehydroneophysalin B, Licoricone, Methyl lucidenate P, Morusimic acid A, Sanjoinenine, Schininallylol
P00734
F2
Coagulation factor II
8
12S-Hydroxyandrographolide,4,7-Didehydroneophysalin B,6-Feruloyl catalpol, Bigelovin, Herbacetin, Heterodendrin, Isoxanthanol, Kaempferol, Licoricone
P08709
F7
Coagulation factor VII
3
Kaempferol, Paristerone, Picrasinoside H
P14867
GABRA1
Gamma-aminobutyric acid receptor subunit alpha-1
4
Herbacetin, Kaempferol, Schininallylol, Zederone
P00390
GSR
Glutathione reductase
2
Maokonine, Morin
P09601
HMOX1
Heme oxygenase 1
2
Isoxanthanol, Kaempferol
P05362
ICAM1
Intercellular adhesion molecule 1
2
Kaempferol, Maglifloenone (Denudatone)
O14920
IKBKB
Inhibitor of nuclear factor kappa-B kinase subunit beta
4
13-Hydroxy-9,11-hexadecadienoic acid, Isoxanthanol, Kaempferol, Morusimic acid B
Q12809
KCNH2
Potassium voltage-gated channel subfamily H member 2
4
4,7-Didehydroneophysalin B, Licoricone, Morusimic acid A, Morusimic acid E
P35968
KDR
Vascular endothelial growth factor receptor 2
9
Asterinin B, Asterinin C, Bufotalin, Catenarin, Coronaric acid, Dibutyl sebacate, Licoricone, Methyl lucidenate P, Ophiopogonanone B
P03956
MMP1
Matrix metalloproteinase-1
9
4,7-Didehydroneophysalin B, Asterinin B, Asterinin C, Asterinin D, Chloranoside A, Isoxanthanol, Kaempferol, Kihadanin B, Lupinifolin
P08253
MMP2
Matrix metalloproteinase-2
9
2′-Hydroxy-4,4′,6′-trimethoxydihydrochalcone,3-(2′-Carboxyphenyl)-4(3H)-quinazolinone,4,7-Didehydroneophysalin B, 6-Gingerol, Coronaric acid, Ganoderenic acid B, Isosamarcandin, Methyl lucidenate P, Picroside III
P14780
MMP9
Matrix metalloproteinase-9
9
3-(2′-Carboxyphenyl)-4(3H)-quinazolinone, 6-Gingerol, Asterinin B, Asterinin C, Asterinin D, Bletilol B, Ganoderenic acid B, Isosamarcandin, Isoxanthanol
P35228
NOS2
Nitric oxide synthase
9
3,7-Dimethyloctane-1,3,6-triol, Campesterol acetate, epi-Kansenone, Ganoderenic acid B, Isoxanthanol, Kaempferol, Licoricone, Maglifloenone (Denudatone), Methyl lucidenate P
P29474
NOS3
Nitric oxide synthase
2
3,7-Dimethyloctane-1,3,6-triol, Kaempferol
Q14432
PDE3A
cGMP-inhibited 3′,5′-cyclic phosphodiesterase A
4
6-Gingerol, Maglifloenone (Denudatone), N-cis-Feruloyl typamine, Schininallylol
P06401
PGR
Progesterone receptor
9
14-Deoxyandrographolide,3,7-Dimethyloctane-1,3,6-triol, Andrograpanin, Bufotalin, epi-Kansenone, Ganoderenic acid B, Isoxanthanol, Kaempferol, Methyl lucidenate P
P37231
PPARG
Peroxisome proliferator-activated receptor gamma
9
1,7-Bis(4-hydroxyphenyl)-hepta-4E,6E-dien-3-one, 13-Hydroxy-9,11-hexadecadienoic acid, Coronaric acid, Herbacetin, Kaempferol, Lupinifolin, Morin, N-Isobutyl-(2E,4E)-octadecadienamide, Saurufuran A
P17612
PRKACA
cAMP-dependent protein kinase catalytic subunit alpha
4
1,7-Bis(4-hydroxyphenyl)-hepta-4E,6E-dien-3-one,2′-Hydroxy-4,4′,6′-trimethoxydihydrochalcone, Indigotin, Kaempferol
P07477
PRSS1
Serine Protease 1
5
6-Feruloyl catalpol, Bigelovin, Herbacetin, Kaempferol, Licoricone
P16581
SELE
Selectin E
2
Kaempferol, Maglifloenone (Denudatone)
P23975
SLC6A2
Solute carrier family 6 member 2
5
1-Methoxy-3,7-dimethyl-2,6-octadiene, 6-Gingerol, epi-Kansenone, Kaempferol, Salsoline
Q01959
SLC6A3
Solute carrier family 6 member 3
8
1-Methoxy-3,7-dimethyl-2,6-octadiene, 6-Gingerol, epi-Kansenone, Isoxanthanol, Lobelanidine, Morusimic acid B, Paristerone, Salsoline
P31645
SLC6A4
Solute carrier family 6 member 4
5
1-Methoxy-3,7-dimethyl-2,6-octadiene, 6-Gingerol, epi-Kansenone, Paristerone, Salsoline
P01375
TNF
Tumour necrosis factor
8
Asterinin B, Asterinin C, epi-Kansenone, Ganoderenic acid B, Kaempferol, Lupinifolin, Methyl lucidenate P, Paristerone
P11387
TOP1
DNA topoisomerase 1
6
Asterinin B, Asterinin C, Chloranoside A, epi-Kansenone, Morin, Picrasinoside H
P47989
XDH
Xanthine dehydrogenase/oxidase
2
Catenarin, Kaempferol
Mol ID1
Compound
PubChem CID2
Molecular Formula
No.3
Target4
OB%5
BBB6
DL7
Structure8
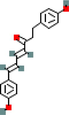
MOL000074
1,7-Bis(4-hydroxyphenyl)-hepta-4E,6E-dien-3-one
10613719
C19H18O3
3
ADRB2, PPARG, PRKACA
67.92
−0.42
0.24
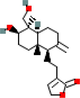
MOL008226
14-Deoxyandrographolide
11624161
C20H30O4
1
PGR
56.3
−0.55
0.31
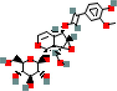
MOL002787
6-Feruloyl catalpol
44566578
C24H30O12
3
CA2, F2, PRSS1
31.38
−2.09
0.84
MOL008219
Andrograpanin
11666871
C20H30O3
1
PGR
56.3
−0.55
0.31
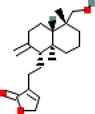
MOL001490
Bis(2-ethylhexyl) Phthalate
8343
C24H38O4
2
ADRB2, CHRM3
43.59
0.68
0.35
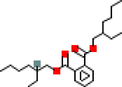
MOL002823
Herbacetin
5280544
C15H10O7
6
ACHE, AR, F2, GABRA1, PPARG, PRSS1
36.07
−0.65
0.27
MOL001781
Indigotin
10215
C16H10N2O2
1
PRKACA
38.2
0.02
0.26
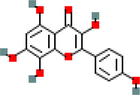
MOL000422
Kaempferol
5280863
C15H10O6
23
ACHE, AKT1, ALOX5, AR, BCL2, CYP1B1, F2, F7, GABRA1, HMOX1, ICAM1, IKBKB, MMP1, NOS2, NOS3, PGR, PPARG, PRKACA, PRSS1, SELE, SLC6A2, TNF, XDH
41.88
−0.55
0.24
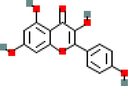
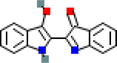
MOL004855
Licoricone
5319013
C22H22O6
7
AR, F10, F2, KCNH2, KDR, NOS2, PRSS1
63.58
−0.14
0.47
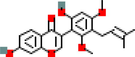
MOL012207
Lobelanidine
442646
C22H29NO2
4
ADRB2, CHRM3, F2, SLC6A3
60.53
0.31
0.32
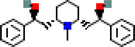
MOL000737
Morin
5281670
C15H10O7
5
ALOX5, AR, GSR, PPARG, TOP1
46.23
−0.77
0.27
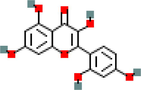
MOL000483
N-cis-Feruloyl Typamine
6440659
C18H19NO4
2
ADRB2, PDE3A
118.35
−0.27
0.26
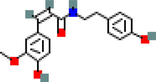
MOL001539
Sanjoinenine
14729078
C29H35N3O4
1
F10
67.28
−0.24
0.79
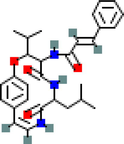
MOL011931
Vitetrifolin E
11143042
C22H36O4
1
PGR
31.41
−0.04
0.3
2.4 Concentration quantification of representative compounds
In order to confirm the presence of chemical components detected via LC-MS, we measure three representative compounds via ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS), which were kaempferol, licoricone ad morin. All experiments were carried out on a Thermo Fisher TSQ LC-MS/MS system (Thermo Fisher Scientific Co., San Jose, CA, USA) consisting of an Accela Autosampler, an Accela pump, and a Quantum Access triple quadrupole mass spectrometer. All system control, data acquisition, mass spectral data evaluation and data processing were performed using XCalibur software version 2.0.7 (Thermo Fischer, San Jose, CA, USA) and Thermo LCquan software version 2.5.6 (Thermo Fischer, San Jose, CA, USA).
The Agilent ZORBAX SB C18 column (2.1 mm × 150 mm, 3.5 μm) was used for separation. The mobile phase consisted of 0.1% formic acid in water (solvent A) and methanol (solvent B). Using a gradient program started with 60% A, a step to 90% A until 8 min, then with a isocratic elution of 90% A until 10 min, and re-equilibration from 10.01 min to 15 min with 60% A at a flow rate of 0.30 mL/min and column temperature at 40 °C. MS conditions were as follows: ESI in negative mode, vaporizer temperature at 280 °C, capillary temperature at 320 °C, sheath gas pressure at 40 psi, auxiliary gas pressure at 20 psi and spray voltage at 3000 V. Quantification was accomplished in multiple reaction monitoring (MRM) by monitoring the transition of m/z 301.2 → 151 for morin, 381.4 → 323 for licoricone and 285.2 → 241 for kaempferol. LC-MS/MS chromatogram with mixed surrogate standards for kaempferol, morin and licoricone was present in Supplementary Figure S3. This developed method was applied for the detection and quantification of the three compounds (Kaempferol, Morin, Licoricone) in the ethanol extract of mulberry leaves with calculated linear regression models (Supplementary Table S3).
2.5 Protein-protein interaction (PPI) network
The 37 identified T2DM targets were imported into the online platform STRING https://string-db.org/, and the organism was set to Homo sapiens (Supplementary Figure S4). High-confidence protein interactions with enrichment P-value<1e-16 were selected, which included 37 nodes and 138 edges with an average node degree of 7.46. A TSV file of PPI network was automatically generated via STRING, which included parameters such as nodes1, node2, and combined_score, etc. (Supplementary Table S4). The file was then input into CytoScape, an open-source software platform for visualizing complex networks https://cytoscape.org/, during which node1 column was set as Source Node, node2 column was set as Target Node, while the remaining columns were set as default parameters. Protein-protein interactions was present in CytoScape as an enrichment network (Fig. 2A). During the analysis, the NetworkAnalyzer plug-in was used to calculate the values of node degrees (Assenov et al., 2008), according to which, the higher the degree value was, the more important the node was in the network. Molecular Complex Detection (Mcode) Clustering ftp://ftp.mshri.on.ca/pub/BIND/Tools/MCODE of T2DM targets in the protein–protein interaction network was then performed with Mcode_Score set to greater than or equal to 6, from which the most highly interconnected regions in a network were identified. According to the analysis, only one sub-network was identified, which was presented in Fig. 2B.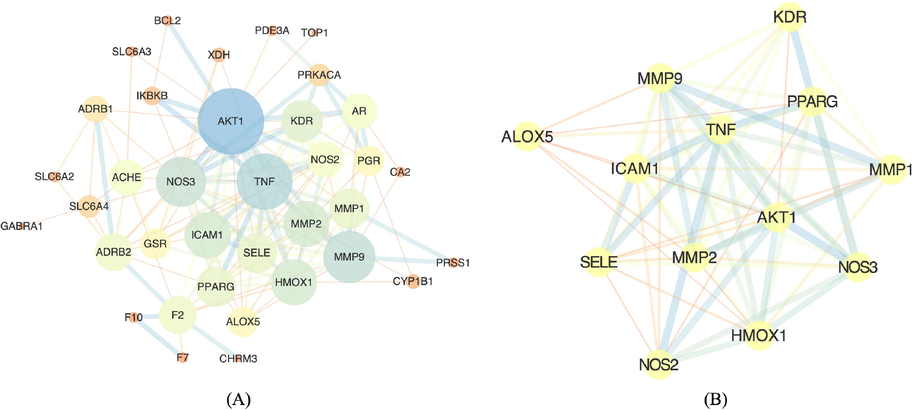
Schematic illustration of the enrichment of protein–protein interactions between target proteins (P-value < 1e−16). Network nodes represent proteins, and the edges represent protein–protein associations. (A) PPI enrichment analysis through online STRING platform version 11.0. (B) Highly inter-connected region through Mcode analysis.
2.6 Compound-target network construction
The interactional relationships between 63 active components in the ethanol extract of mulberry leaves and 37 corresponding targets were imported into CytoScape. Then, the active components and target genes were drawn into circle by using the function of Degree Circle Layout within the plug-in Layout. Moreover, components collected from TCMSP were coloured in blue while those predicted by PharmMapper were coloured in red. In addition, all target genes were in green colour. NetworkAnalyzer function in the plug-in Tools was used to generate the degree values of each node in the entire network graph, and then the Generate Style from Statistics function was used to draw the component-target network graph based on the degree values. That is, the sizes (diameters) of the nodes in the network were determined by degree values. The greater the degree value is, the larger the node diameter is, and the more important the node is in the whole network.
2.7 Gene ontology (GO) enrichment analysis
In order to clarify the functions of target compounds in the ethanol extract of mulberry leaves and their role in signal transduction, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) to carry out GO functional enrichment of all the identified target proteins. According to the analysis, molecular function (MF), cellular component (CC) and biological process (BP) of target proteins were respectively described. GO analysis also returned a P-value for each GO term with a small P-value indicating that most different genes were enriched. According to the hypergeometric distribution relationship, a small P-value (<0.05) indicates enrichment of differential gene in the GO analysis. In addition, the smaller the P-value, the more significant the differential enrichment is. Since small P-value is not well present, we performed the -log10 (P-value) conversion. Thus, the larger the -log10 (P-value), the more significant the differential enrichment is, which is convenient for intuitive understanding of the meaning of the data during visualization.
2.8 KEGG pathway enrichment analysis
Target genes were imported into the Ensemble database (http://asia.ensembl.org/index.html). BioMart function in Ensemble was then used to convert the gene IDs into Entrez Gene IDs, which were entered into KOBAS (http://kobas.cbi.pku.edu.cn/kobas3/?T=1). Gene-List Enrichment function in KOBAS was used, Species was set to Homo Sapiens, Input Type was set to Entrez Gene ID, Input was set to Intersection Genes, while Database was selected as KEGG Pathway (K) in PATHWAY. To further verify that whether the biological processes related to the target protein are associated with the type 2 diabetes mellitus, a total of 30 pathways filtered with P-value < 0.05 were selected for further KEGG pathway analysis while the corresponding -log10(P-value) of each pathway was visualized. After the analysis of KEGG pathway enrichment, both bar plot and bubble plot were generated. It was noteworthy that -log10(P-value) had the same meaning as stated in Section 2.7.
2.9 Target pathway analysis
KEGG Mapper Tool was used to construct the pathway mapping in terms of the ethanol extract of mulberry leaves for the treatment of type 2 diabetes mellitus. The pathway map showed that ethanol extract of mulberry leaves was involved in the treatment of diabetes through pathways such as PI3K-AKT signaling pathway, NF-κB signaling pathway, and MAPK signaling pathway, etc.
3 Results
3.1 Identification of anti-diabetic components and target genes
According to the LC-MS analysis, a total of 248 components were identified in the ethanol extract of mulberry leaves, which were all searched in the TCMSP database and a total of 86 compounds were found (Ru et al., 2014). Through using OB ≥ 30% and DL ≥ 0.18 as criteria for filtering, 18 compounds were selected, which were considered as active components and were linked with 96 target genes. As for other compounds (n = 162) that were not found in TCMSP database, we used PubChem to generate their two-dimensional structures (Kim et al., 2016). These structures were then imported into the PharmMapper server to predict whether they were associated with any disease (Liu et al., 2010). A total of 58 compounds were identified to be associated with 633 diseases via 328 target proteins, the names of which were then mapped through the Gene-Disease Association Database to get the names of target genes in TCM-Mesh server (Zhang et al., 2017). Moreover, the two databases, GeneCards and OMIM, were thoroughly searched for target genes that have been linked with diabetes by using the keyword diabetes-type 2. A total of 9948 target genes were collected. In order to find out the most relevant genes associated with both type 2 diabetes and components of mulberry leaves, the intersection of targets from TCMSP, PharmMapper, and GeneCards/OMIM were obtained. A total of 37 targets were identified, the details of which were recorded in Table 1. UniProt ID, gene names, protein names, and the interacted compounds of the targets were all included in the table.
Further analysis showed that the 37 targets interacted with 63 compounds of mulberry leaves. 14 compounds were identified from TCMSP with confirmed biological activity toward T2DM and their PubChem CID, molecular formula, oral bio-availability, blood–brain barrier, drug likeness, and 2-Dstructure were all present in Table 2. Among the 14 bioactive compounds, we quantified the concentration of three representative chemical components (Kaempferol, Licoricone and morin) via LC-MS/MS, according to which, content of morin was 1.58 ± 0.05 mg/g, kaempferol 7.28 ± 0.12 mg/g, and licoricone 0.31 ± 0.02 mg/g. The results were obtained with repetition in triplicates. As for the other 49 compounds, they were potentially bioactive in T2DM treatment and their information was collected from PharmMapper. The details of these compounds were recorded in Supplementary Table S2.
3.2 Network of target protein–protein interactions
All target proteins were imported into the online database STRING https://string-db.org/ version 11.0 to construct the protein–protein interaction network (Szklarczyk et al., 2019). According to the result, the network has significantly more interactions (n = 138) than expected (n = 41), which means that these proteins have more functional and physical interactions among themselves and could be biologically connected as a group (Supplementary Figure S4). In order to better illustrate the interactions between proteins, the network was then visualized in CytoScape (Fig. 2A), in which larger nodes means greater degrees (more connections with other nodes), hence pivotal roles in the network. As for the line thickness, it is calculated based on the combined score generated through STRING database. The thicker the line is, the stronger the connections are between the two nodes. Mcode clustering of the network in Fig. 2A was performed, from which the most highly inter-connected regions in a network were identified (Fig. 2B). According to the results, AKT1 is the core target gene in the network while 13 target genes (AKT1, TNF, MMP9, ICAM1, NOS3, MMP2, KDR, SELE, NMP1, NOS2, HMOXX1, PPARG, and ALOX5) were tightly linked together.
3.3 Interactions between anti-diabetic compounds and target genes
Generally speaking, a target can be modulated by multiple compounds while a compound could act on multiple targets. To better understand the interactions between 63 anti-diabetic compounds and 37 target genes, we constructed a network by using CytoScape 3.8.2, which included 100 nodes and 233 edges (Fig. 3). In the network, the degree of a node represents its connections with other nodes, which was reflected by the diameter of the node. That is, the nodes with more connections play pivotal roles in the entire interaction network, which indicates that the nodes could be key target genes during the treatment of type 2 diabetes or compounds that play important anti-diabetic roles in the ethanol extract of mulberry leaves. In specificity, the green circles represent 37 target genes (proteins), the blue circles represent 14 compounds collected from TCMSP database, while the pink circles represent 49 compounds predicted in the PharmMapper database. Grey lines connect circles together, indicating the interactions between the components and the targets. In addition, according to the diameters of the nodes, kaempferol (n = 23) and methyl lucidenate P (n = 11) are the two key important compounds from TCMSP and PharmMapper that have the highest number of interactions with target genes, respectively. In contrast, multiple target genes were modulated by the same number of compounds, which included acetylcholinesterase, polyunsaturated fatty acid 5-lipoxygenase, androgen receptor, vascular endothelial growth factor receptor 2, interstitial collagenase, 72 kDa type IV collagenase, matrix metalloproteinase 9, nitric oxide synthase, MORF4 family-associated protein 1, and peroxisome proliferator-activated receptor gamma.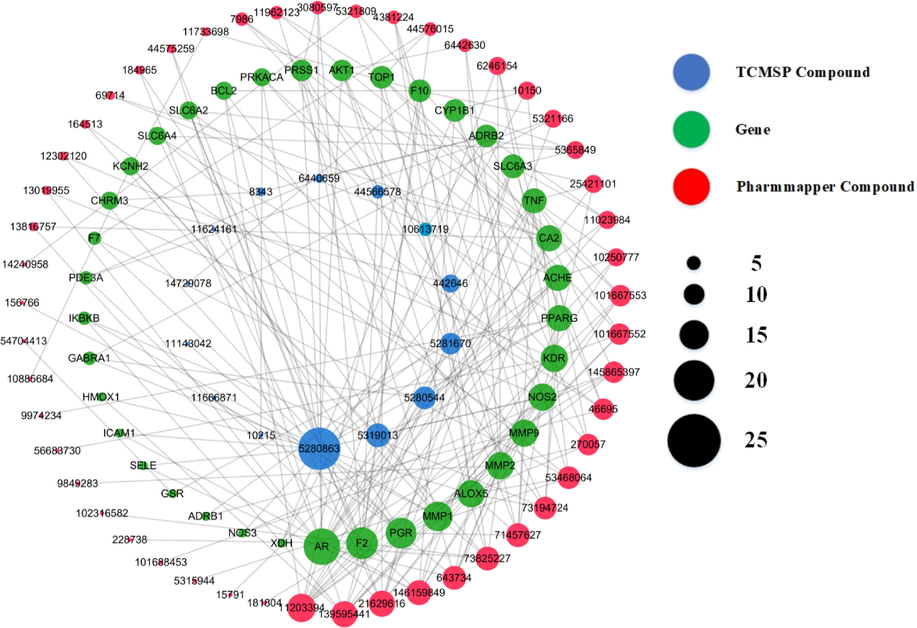
The interaction network among components from the ethanol extract of mulberry leaves and T2DM-related target proteins. All the compounds were labelled with PubChem ID in order to avoid compound-name overlapping. The diameters of circles reflect the number of interactions. Green circles: target genes (proteins). Blue circles: 14 compounds collected from TCMSP database. Pink circles: 49 compounds collected from PharmMapper database. Grey lines: connections between circles.
3.4 GO and KEGG pathway enrichment analysis
GO describes gene products with three independent categories, which include biological process, cellular component, and molecular function (Consortium, 2019). GO enrichment analysis was performed in order to find out which GO terms are over- or under-represented by using annotations for a specific gene set. In this study, 37 target proteins were analysed through DAVID (Dennis et al., 2003) and top 10 GO entries for each category were selected according to false discovery rate (FDR, < 0.05) as shown in Fig. 4.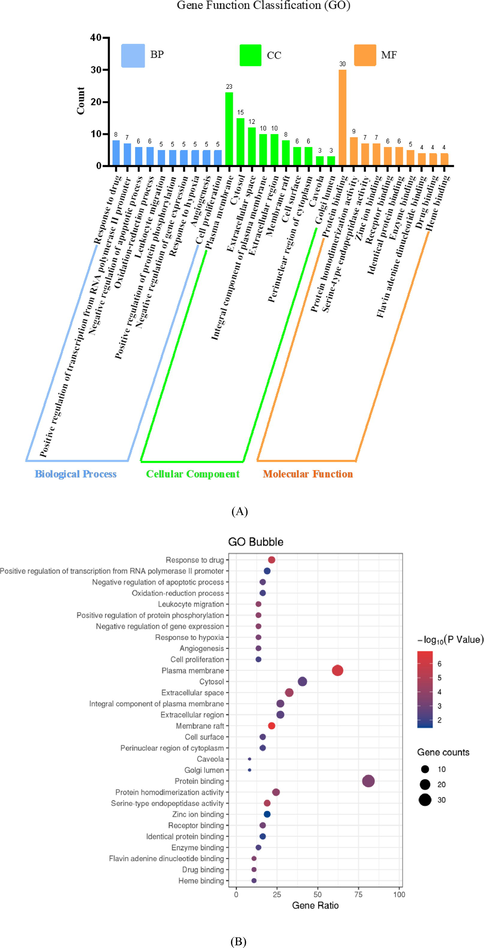
GO enrichment analysis of target proteins. (A) Bar plot of the number of GO entries in the functional categories of biological process (BP, blue bar), cell composition (CC, green bar), and molecular function (MF, orange bar). (B) Bubble plot of GO entries filtered with -log10(P-value < 0.05).
A total of 30 pathways filtered from KEGG analysis with P-value < 0.05 were selected and visualized as bar plot with the number of target proteins in each of the KEGG pathways (Fig. 5A) while the corresponding -log10(P-value) of each pathway was shown as bubble plot in Fig. 5B. Various inflammatory signaling pathways such as TNF signaling pathway, calcium signaling pathway, cGMP-PKG signaling pathway, cAMP signaling pathway, regulation of lipolysis in adipocytes, NF-κB signaling pathway, insulin resistance pathway, and sphingolipid signaling pathway were associated with type 2 diabetes mellitus. Thus, according to the GO and KEGG pathway enrichment analyses, components from the ethanol extract of mulberry leaves were involved in a variety of signaling, signal transduction, and lipolysis pathways associated with T2DM.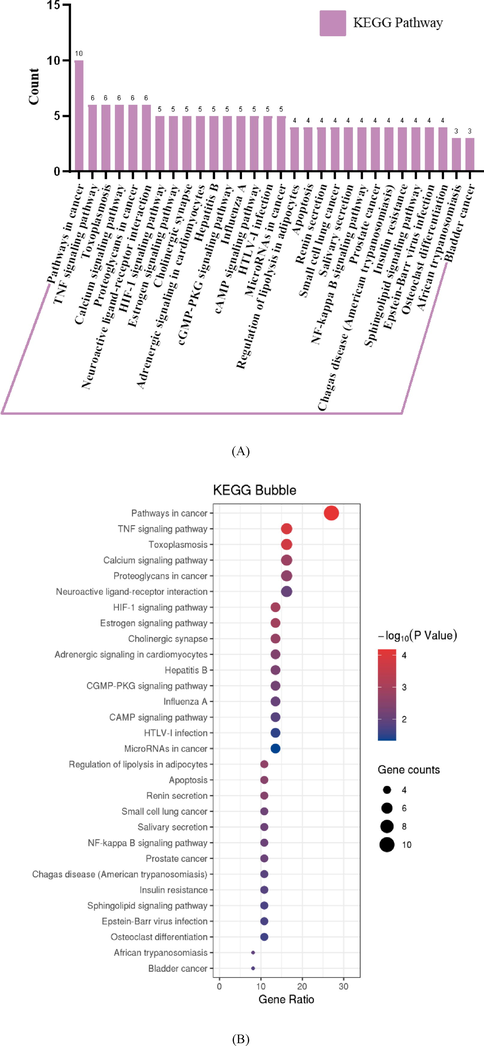
Target protein KEGG pathway analysis. (A) Bar plot of the number of target proteins in each of the KEGG pathways. (B) Bubble plot of KEGG pathways. All the KEGG pathways were filtered with P-value < 0.05.
In order to elucidate the interactions of target proteins with all the identified KEGG pathways, CytoScape was used to construct a target-pathway network (Fig. 6). According to the diagram, the target proteins involved in the TNF signaling pathway included inhibitor of nuclear factor kappa-B kinase subunit beta (IKBKB), RAC-alpha serine/threonine-protein kinase (AKT1), E-selectin (SELE), tumour necrosis factor (TNF), matrix metalloproteinase 9 (MMP9), and intercellular adhesion molecule 1 (ICAM1). The target proteins involved in the calcium signaling pathway included muscarinic acetylcholine receptor M3 (CHRM3), nitric oxide synthase 2 (NOS2), nitric oxide synthase 3 (NOS3), Beta-1 adrenergic receptor (ADRB1), Beta-2 adrenergic receptor (ADRB2), and cAMP-dependent protein kinase catalytic subunit alpha (PRKACA). The target proteins involved in the cGMP-PKG signaling pathway included NOS3, cGMP-inhibited 3′ (PDE3A), AKT1, ADRB1, and ADRB2. The target proteins involved in the cAMP signaling pathway included PDE3A, AKT1, ADRB1, ADRB2, and PRKACA. The target proteins involved in the regulation of lipolysis in adipocytes included AKT1, ADRB1, ADRB2, and PRKACA. The target proteins involved in the NF-κB signaling pathway included IKBKB, apoptosis regulator Bcl-2 (BCL2), TNF, and ICAM1. The target proteins involved in the insulin resistance pathway included IKBKB, NOS3, AKT1, and TNF. The target proteins involved in the sphingolipid signaling pathway included NOS3, BCL2, AKT1, and TNF.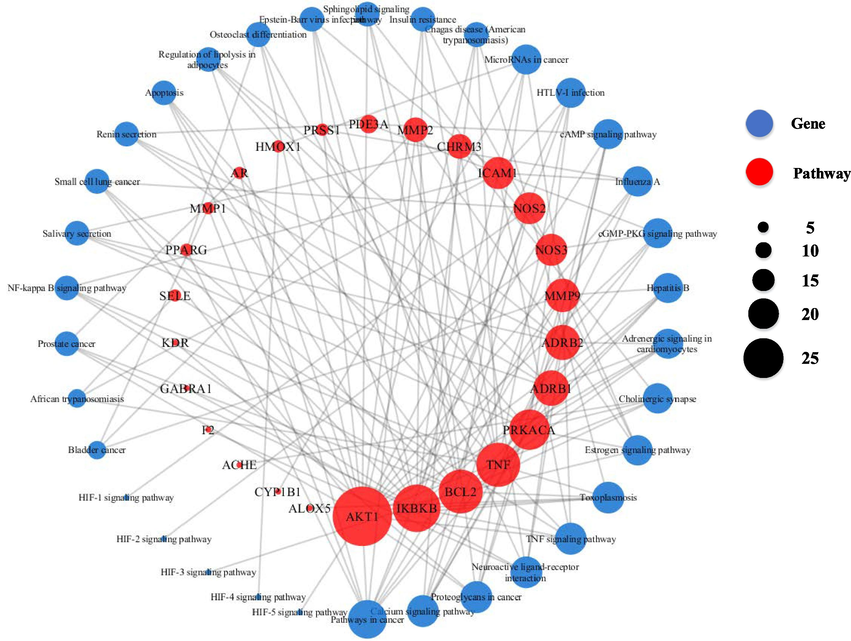
Schematic illustration of the network of pathway-target interactions. Blue-circle nodes represent the pathways involved in the target protein while pink-circle nodes represent target proteins. The diameter of the circle node represents the number of connections (interactions) with other nodes.
3.5 Target path analysis
The 37 selected targeted genes were submitted to the Ensemble database by BioMart, through which the gene names were converted to Entrez Gene ID. KOBAS database was then used for KEGG pathway analysis, and target genes in the given analysis results were arranged in ascending order based on the P-values. The statistically significant target genes in pathways were highlighted in red and present in Fig. 7. The pathway map showed that ethanol extract of mulberry leaves was involved in the treatment of diabetes through regulating specific pathways, including PI3K-AKT signaling pathway, NF-κB signaling pathway, and MAPK signaling pathway, etc.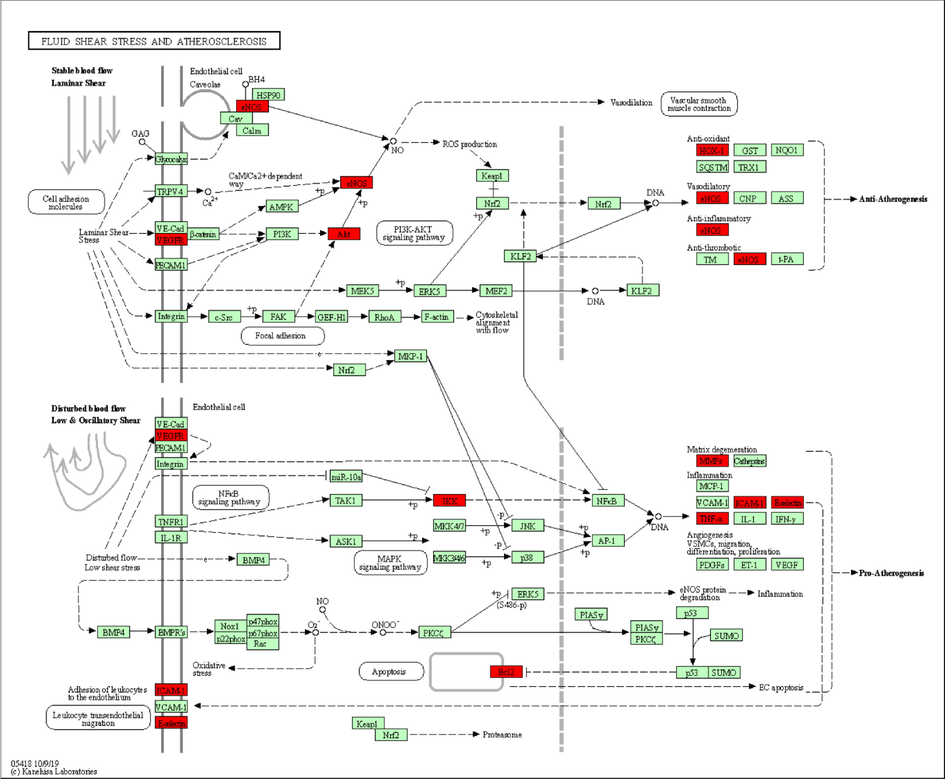
Pathway map of ethanol extract of mulberry leaves in the treatment of type 2 diabetes mellitus. The main target proteins that are involved in diabetes-related pathways are highlighted in red. Arrows represent the activation effect, T-arrows represent the inhibition effect, and segments show the activation effect or inhibition effect.
4 Discussion
The philosophy of traditional Chinese medicine has been considering a patient as an integrative system. With the long-term development of TCM, it has many effective herbal formulae to treat a variety of chronic diseases such as nephritis and diabetes, etc. (Li and Zhang, 2013). However, due to the complexity of TCMs, it is rather difficult to elucidate their molecular mechanisms. For the past twenty years, network pharmacology, as an interdisciplinary science through integration of pharmacology, bioinformatics, and other related scientific disciplines, has been widely used in the discovery of novel disease targets and active drug compounds (Hopkins, 2008). Since the essential idea of network pharmacology focuses on the “multi-target, multi-drug” paradigm, it intrinsically matches with the principle of traditional Chinese medicine (Li and Zhang, 2013). Mulberry leaves have been used as an effective TCM to combat inflammation and reduce hypertension for a long time in Asian countries (Thaipitakwong et al., 2018). In addition, many studies reported the beneficial effects of mulberry leaves in lowering blood glucose level and alleviating the conditions of diabetic diseases (He et al., 2019; Li et al., 2019; Tian et al., 2019; Li et al., 2020). Recently, some studies have used network pharmacology to investigate the molecular mechanisms of mulberry leaves in treating diabetes (Ge et al., 2018; Wu and Hu, 2020). However, only components sourced from public database or volatile components in the mulberry leaves were examined.
In this study, we explored the component compositions of ethanol extract of mulberry leaves via LC-MS, which was then systematically searched in a variety of public databases in order to identify potentially effective components, target genes, and important pathways for the treatment of T2DM. According to the result, a total of 63 compounds and 37 target proteins were identified (Table 1), among which 14 compounds sourced from TCMSP database were biologically active (Table 2). As for the 37 target proteins, PPI analysis revealed that all of them formed an integrative network except for one target potassium voltage-gated channel subfamily H member 2 (KCNH2). It was previously reported that the mutations of KCNH2 gene could cause the rare syndrome congenital long QT syndrome (LQTS) that carries an increased risk of cardiac arrhythmias (Oshiro et al., 2010) while a recent study revealed LQTS patients with KCNH2 mutations display an increased insulin release with an increased risk of hypoglycaemia (Marstrand et al., 2019). Clusters in the PPI network are normally protein complexes or belong to the same pathway. Compared with the non-interacting protein pairs, we found that the predicted PPIs tend to be in the same or related pathways (PPI enrichment P-value < 1.0e-16), which indicated that our predicted PPIs were reliable.
Further Mcode analysis of the PPIs revealed that 13 targets were highly inter-connected, which included AKT1, ALOX5, HMOX1, ICAM1, KDR, MMP1, MMP2, MMP9, NOS2, NOS3, PPARG, SELE, and TNF. AKTs have three different isoforms (AKT1, AKT2, and AKT3), in which AKT1 expresses ubiquitously while its blocking in pancreatic β-cells and peripheral tissues lead to hyperglycaemia and diabetes (Huang et al., 2018). As for ALOX5 gene, it encodes 5-lipoxygenase and is implicated in chronic inflammatory disease, the polymorphism of which has also been identified to confer genetic susceptibility to type 2 diabetes (Nejatian et al., 2019). HMOX1 is the coding gene of heme oxygenase 1 that is essential for haem degradation, a risk factor in the progression of type 2 diabetes and obesity-associated metabolic disturbances (Moreno-Navarrete et al., 2017). Intercellular adhesion molecule 1 (ICAM1) is a cell surface glycoprotein express, both genetic polymorphism and gene expression level of which are correlated with diabetes and diabetic nephropathy (Gu et al., 2013). As for KDR, it encodes vascular endothelial growth factor receptor 2, which plays important roles in mediating vasculogenic and angiogenic processes that involves diabetic retinopathy (Sassa et al., 2004). Collagenases (MMP1) and gelatinases (MMP2 and MMP9) belong to the protein group of matrix metalloproteinases and function as zinc-binding proteolytic enzymes, increased activities of which are responsible for a variety of diseases such as tumour growth, arthritis and cardiovascular disease (Lewandowski et al., 2011). In addition, it was also revealed that MPPs were involved in tissue remodelling, maintaining an integral role in diabetic wound healing (Ayuk et al., 2016). NOS2 gene encodes the inducible nitric oxide synthase while NOS3 genes encodes endothelial nitric oxide synthase gene, both of which have been proved to play important roles in the susceptibility to type 2 diabetes mellitus (T2DM) and diabetic nephropathy (Chen et al., 2016). Peroxisome proliferator-activated receptor gamma (PPARG) could increase the systemic insulin sensitization through complex mechanisms involving multiple pathways when activated (Lv et al., 2017). As for SELE, it encodes endothelial adhesion molecule 1 and its increased concentration was associated with both insulin-dependent and non-insulin-dependent diabetes mellitus patients (Cominacini et al., 1995). In terms of TNF, it is a tumour necrosis factor that acts as one of the most important pro-inflammatory mediators in the development of insulin resistance and pathogenesis of T2DM (Akash et al., 2018).
In order to understand the interactions among compounds and targets, we constructed a multi-layer network, which provided insights into compound-target relationships (Fig. 3). According to the results, 14 active compounds from the ethanol extract of mulberry leaves were found in TCMSP database. Among them, three representative compounds were quantitatively measured in terms of their concentrations, including Kaempferol (n = 23), Licoricone (n = 7), Morin (n = 5), all of which were interplayed with multiple targets related with T2DM. For example, the natural flavanol kaempferol has the highest number of connections with target proteins. Thus, it indicates that kaempferol may be a key component in the ethanol extract of mulberry leaves and plays important roles in the treatment of diabetes. In fact, many studies have reported the anti-diabetic effects of the compound. In particular, An et al. (2011) studied kaempferol extracted from Ginkgo biloba and found that it could reduce hyper-glycemia syndrome and enhance glucose uptake through mimicking the action of insulin. Ishaq et al. (2019) summarized the anti-diabetic effect of kaempferol, according to which the compound could systematically alleviate diabetic conditions such as increasing production and secretion of insulin, enhancing synthesis of new glucose transporters, and suppressing hepatic gluconeogenesis, etc. Morin is another natural dietary flavonoid compound that has been found in different herbs, fruits and wine (Gallyas et al., 2012). According to Paoli et al. (2013), morin could inhibit gluconeogenesis and enhances glycogen synthesis, which has the potential to be developed into an low-molecular-weight antidiabetic drug. Another study by Lin et al. (2017) revealed that morin could decrease blood glucose level by increasing plasma insulin secretion in diabetic rats by acting as an agonist of imidazoline I-3 receptor, which also indicated that develop morin had the potential as a drug for the treatment of diabetic disorders. Multiple studies also reported the anti-diabetic functions of other bioactive compounds in the ethanol extract of mulberry leaves, which validated the methodology of network pharmacology used in this study for the discovery of the interactions between compounds, targets and type 2 diabetes mellitus. However, it was noteworthy that, although licoricone was identified as a meaningful compound to combat T2DM, there were very rare experimental studies confirming its anti-diabetic roles, which indicated that more efforts should be devoted to this drug in the study of T2DM.
As for the 49 compounds identified through PharmMapper prediction, compounds such as methyl lucidenate P, isoxanthanol, and epi-Kansenone diabetes, etc. have functions in the treatment of diabetic pathogenesis. Methyl lucidenate P has impacts on 11 target proteins while epi-Kansenone interacts with 9 target proteins. However, literature analysis showed that there are few reports of these compounds in diabetic treatment, which suggested that these compounds were worthy of further investigations in terms of their anti-diabetic functions. As for the compound-target interactions, ACHE (acetylcholinesterase), ALOX5 (polyunsaturated fatty acid 5-lipoxygenase), AR (androgen receptor), KDR (vascular endothelial growth factor receptor 2), MMP1 (matrix metalloproteinase-1), MMP2 (matrix metalloproteinase-2), MMP9 (matrix metalloproteinase-9), NOS2 (nitric oxide synthase), PGR (progesterone receptor), and PPARG (peroxisome proliferator-activated receptor gamma) are among the highly connected proteins with compounds, all of which have 9 connections. For example, androgen receptor with reduced insulin sensitivity is associated with poor glycaemic control in men with type 2 diabetes while acetylcholinesterase is associated with the apoptosis of β cells, which contributes to the pathogenesis of insulin-dependent diabetes mellitus (Zhang et al., 2012).
According to the KEGG pathway analysis, protein targets regulated by the ethanol extract of mulberry leaves were involved in pathways such as pathways in cancer, TNF signaling pathway, NF-κB signaling pathway, and insulin resistance pathway, etc. It has been reported that there is a cross-talk between the multiple pathways at the interface of the diabetes–cancer link, which may explain why target proteins for diabetes fall in to the pathways in cancer (Tudzarova et al., 2015). In TNF signaling pathway, TNF-α is considered as a causative factor in insulin resistance and the pathogenesis of type 2 diabetes (Moller, 2000), which is also interacted with insulin signaling pathway through mechanisms such as phosphorylation of IRS-1 (insulin receptor substrate 1) (Alipourfard et al., 2019). As for NF-κβ signaling pathway, NF-κβ is a potential target for the vascular complications in diabetes because hyperglycaemia activates NF-κβ, leading to the increased expression of cytokines, chemokines and cell adhesion molecules (Suryavanshi and Kulkarni, 2017). In the target-pathway analysis (Fig. 6), RAC-alpha serine/threonine-protein kinase (AKT1) was identified as the most important target that was present in 22 different pathways. AKT belongs to the PI3K/AKT signalling pathway that is required for normal metabolism of glucose, the imbalance of which could lead to the development of type 2 diabetes mellitus (Huang et al., 2018). It was followed by inhibitor of nuclear factor kappa-B kinase subunit beta (IKBKB) and apoptosis regulator Bcl-2 (BCL2), which were involved in 15 and 13 pathways, respectively. The metabolic disorders of insulin resistance and type 2 diabetes are normally associated with chronic inflammations while IKBKB encoding IKK-β is a central coordinator of inflammatory responses that is correlated with the activation of NF-κβ (Arkan et al., 2005). In terms of BCL2, it is essential in controlling the mitochondrial pathway of β-cell apoptosis (Gurzov and Eizirik, 2011). In addition, it is also important in regulating glucose metabolism (Gurzov and Eizirik, 2011). Thus, BCL2 is closely associated with the pathogenesis of type 2 diabetes mellitus.
5 Conclusion
This study systematically examined the effects of chemical components identified in the ethanol extract of mulberry leaves via LC-MS on the treatment of type 2 diabetes mellitus. A total of 14 active components and 49 potentially functional compounds were identified from TCMSP and PharmMapper database, respectively, which worked on 37 target proteins directly involved in the pathogenesis of T2DM. Kaempferol was shown to be the most influential active compound while androgen receptor was most widely regulated by compounds in the ethanol extract of mulberry leaves. Potentially effective components were also identified through compound-target network analysis. Protein-protein interaction and target-pathway network revealed that AKT1 was most important in the 37 target proteins in terms of its functions in different pathways and interactions with other target proteins. Moreover, pathway analysis showed that ethanol extract of mulberry leaves alleviated type 2 diabetes mellitus through pathways such as TNF signaling pathway, NF-κB signaling pathway, and insulin resistance pathway, etc. In sum, network pharmacology provided a systematic view of the functions of the ethanol extract of mulberry leaves in the treatment of type 2 diabetes mellitus, which theoretically validated the effects of mulberry leaves at molecular level. In addition, candidate target proteins and compounds were identified for further experimental studies in terms of their roles in diabetic pathogenesis and treatment.
Funding
Professor Liang Wang would like to thank the financial support of National Natural Science Foundation of China (No.31900022, No.32171281), Natural Science Foundation of Jiangsu Province (No.BK20180997), Young Science and Technology Innovation Team of Xuzhou Medical University (No.TD202001) and Jiang-Su Qing-Lan Project (2020).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Tumor necrosis factor-alpha: role in development of insulin resistance and pathogenesis of type 2 diabetes mellitus. J. Cell. Biochem.. 2018;119(1):105-110.
- [Google Scholar]
- TNF-α downregulation modifies insulin receptor substrate 1 (IRS-1) in metabolic signaling of diabetic insulin-resistant hepatocytes. Mediators Inflamm.. 2019;2019:1-6.
- [Google Scholar]
- The bioflavonoid kaempferol Is an Abcg2 substrate and inhibits Abcg2-mediated quercetin efflux. Drug Metab. Dispos.. 2011;39(3):426-432.
- [Google Scholar]
- IKK-β links inflammation to obesity-induced insulin resistance. Nat. Med.. 2005;11(2):191-198.
- [Google Scholar]
- Assenov, Y., Ramírez, F., Schelhorn, S.-E., Lengauer, T., and Albrecht, M. (2008). Computing topological parameters of biological networks. Bioinformatics 24, 282-284.
- The role of matrix metalloproteinases in diabetic wound healing in relation to photobiomodulation. J. Diabet. Res.. 2016;2016:1-9.
- [Google Scholar]
- Association of NOS2 and NOS3 gene polymorphisms with susceptibility to type 2 diabetes mellitus and diabetic nephropathy in the Chinese Han population. IUBMB Life. 2016;68(7):516-525.
- [Google Scholar]
- Elevated levels of soluble E-selectin in patients with IDDM and NIDDM: relation to metabolic control. Diabetologia. 1995;38(9):1122-1124.
- [Google Scholar]
- Consortium, T.G.O. (2019). The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Research 47, D330-D338.
- Dennis, G., Sherman, B.T., Hosack, D.A., Yang, J., Gao, W., Lane, H.C., and Lempicki, R.A., 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology 4.
- Protective role of morin, a flavonoid, against high glucose induced oxidative stress mediated apoptosis in primary rat hepatocytes. PLoS ONE. 2012;7
- [Google Scholar]
- Analysis of mulberry leaf components in the treatment of diabetes using network pharmacology. Eur. J. Pharmacol.. 2018;833:50-62.
- [Google Scholar]
- Gu, H.F., Ma, J., Gu, K.T., Brismar, K., 2013. Association of intercellular adhesion molecule 1 (ICAM1) with diabetes and diabetic nephropathy. Front. Endocrinol. 3.
- Bcl-2 proteins in diabetes: mitochondrial pathways of β-cell death and dysfunction. Trends Cell Biol.. 2011;21(7):424-431.
- [Google Scholar]
- He, X., Li, H., Gao, R., Zhang, C., Liang, F., Sheng, Y., Zheng, S., Xu, J., Xu, W., Huang, K., 2019. Mulberry leaf aqueous extract ameliorates blood glucose and enhances energy expenditure in obese C57BL/6J mice. J. Functional Foods 63.
- Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol.. 2008;4(11):682-690.
- [Google Scholar]
- The PI3K/AKT pathway in obesity and type 2 diabetes. Int. J. Biol. Sci.. 2018;14(11):1483-1496.
- [Google Scholar]
- Ishaq, A.L., Abotaleb, Kubatka, Kajo, Büsselberg, 2019. Flavonoids and their anti-diabetic effects: cellular mechanisms and effects to improve blood sugar levels. Biomolecules 9.
- PubChem substance and compound databases. Nucleic Acids Res.. 2016;44(D1):D1202-D1213.
- [Google Scholar]
- Matrix metalloproteinases in type 2 diabetes and non-diabetic controls: effects of short-term and chronic hyperglycaemia. Arch. Med. Sci.. 2011;2:294-303.
- [Google Scholar]
- Li, Q., Liu, F., Liu, J., Liao, S., Zou, Y., 2019. Mulberry Leaf Polyphenols and Fiber Induce Synergistic Antiobesity and Display a Modulation Effect on Gut Microbiota and Metabolites. Nutrients 11.
- Mulberry leaf polyphenols attenuated postprandial glucose absorption via inhibition of disaccharidases activity and glucose transport in Caco-2 cells. Food Funct.. 2020;11(2):1835-1844.
- [Google Scholar]
- Traditional Chinese medicine network pharmacology: theory, methodology and application. Chinese J. Nat. Med.. 2013;11(2):110-120.
- [Google Scholar]
- Identification of morin as an agonist of imidazoline I-3 receptor for insulin secretion in diabetic rats. Naunyn-Schmiedeberg's Arch. Pharmacol.. 2017;390(10):997-1003.
- [Google Scholar]
- Liu, X., Ouyang, S., Yu, B., Liu, Y., Huang, K., Gong, J., Zheng, S., Li, Z., Li, H., and Jiang, H. (2010). PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucl. Acids Res. 38, W609-W614.
- Lv, X., Zhang, L., Sun, J., Cai, Z., Gu, Q., Zhang, R., Shan, A., 2017. Interaction between peroxisome proliferator-activated receptor gamma polymorphism and obesity on type 2 diabetes in a Chinese Han population. Diabetol. Metabolic Syndrome 9.
- Marstrand, P., Theilade, J., Andersson, C., Bundgaard, H., Weeke, P.E., Tfelt-Hansen, J., Jespersen, C., Gislason, G., Torp-Pedersen, C., Kanters, J.K., and Jørgensen, M.E., 2019. Long QT syndrome is associated with an increased burden of diabetes, psychiatric and neurological comorbidities: a nationwide cohort study. Open Heart 6.
- Potential role of TNF-α in the pathogenesis of insulin resistance and Type 2 diabetes. Trends Endocrinol. Metab.. 2000;11(6):212-217.
- [Google Scholar]
- HMOX1 as a marker of iron excess-induced adipose tissue dysfunction, affecting glucose uptake and respiratory capacity in human adipocytes. Diabetologia. 2017;60(5):915-926.
- [Google Scholar]
- 5-Lipoxygenase (ALOX5): Genetic susceptibility to type 2 diabetes and vitamin D effects on monocytes. J. Steroid Biochem. Mol. Biol.. 2019;187:52-57.
- [Google Scholar]
- Oshiro, C., Thorn, C.F., Roden, D.M., Klein, T.E., Altman, R.B., 2010. KCNH2 pharmacogenomics summary. Pharmacogenet. Genom.
- The insulin-mimetic effect of Morin: A promising molecule in diabetes treatment. Biochimica et Biophysica Acta (BBA) - General Subjects. 2013;1830(4):3102-3111.
- [Google Scholar]
- Impact of mulberry leaf extract on type 2 diabetes (Mul-DM): a randomized, placebo-controlled pilot study. Complement. Therap. Medi.. 2017;32:105-108.
- [Google Scholar]
- Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., Li, P., Guo, Z., Tao, W., Yang, Y., Xu, X., Li, Y., Wang, Y., Yang, L., 2014. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics 6.
- Safran, M., Dalah, I., Alexander, J., Rosen, N., Iny Stein, T., Shmoish, M., Nativ, N., Bahir, I., Doniger, T., Krug, H., Sirota-Madi, A., Olender, T., Golan, Y., Stelzer, G., Harel, A., Lancet, D, 2010. GeneCards Version 3: the human gene integrator. Database 2010, baq020-baq020.
- Bifunctional properties of peroxisome proliferator-activated receptor 1 in KDR gene regulation mediated via interaction with both Sp1 and Sp3. Diabetes. 2004;53:1222-1229.
- [Google Scholar]
- Suryavanshi, S.V., Kulkarni, Y.A., 2017. NF-κβ: A Potential Target in the Management of Vascular Complications of Diabetes. Frontiers in Pharmacology 8.
- Szklarczyk, D., Gable, A.L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., Simonovic, M., Doncheva, N.T., Morris, J.H., Bork, P., Jensen, L.J., Mering, Christian v., 2019. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucl. Acids Res. 47, D607-D613.
- Mulberry leaves and their potential effects against cardiometabolic risks: a review of chemical compositions, biological properties and clinical efficacy. Pharm. Biol.. 2018;56(1):109-118.
- [Google Scholar]
- Tian, S., Wang, M., Liu, C., Zhao, H., Zhao, B., 2019. Mulberry leaf reduces inflammation and insulin resistance in type 2 diabetic mice by TLRs and insulin Signalling pathway. BMC Complement. Alternat. Med. 19.
- The role of inflammation in diabetes: current concepts and future perspectives. Eur. Cardiol. Rev.. 2019;14(1):50-59.
- [Google Scholar]
- The double trouble of metabolic diseases: the diabetes–cancer link. Mol. Biol. Cell. 2015;26(18):3129-3139.
- [Google Scholar]
- Treating type 2 diabetes mellitus with traditional Chinese and Indian medicinal herbs. Evid.-Based Complement. Alternat. Med.. 2013;2013:1-17.
- [Google Scholar]
- Systematic evaluation of the mechanisms of Mulberry leaf (Morus alba Linne) acting on diabetes based on network pharmacology and molecular docking. Comb. Chem. High Throughput Screening. 2021;24(5):668-682.
- [Google Scholar]
- Yu, A., Adelson, D., Mills, D., 2018. Chinese Herbal Medicine Versus Other Interventions in the Treatment of Type 2 Diabetes. J. Evid.-Based Integrat. Med. 23.
- Acetylcholinesterase is associated with apoptosis in cells and contributes to insulin-dependent diabetes mellitus pathogenesis. Acta Biochim. Biophy. Sin.. 2012;44:207-216.
- [Google Scholar]
- Zhang, R., Zhu, X., Bai, H., Ning, K., 2019. Network pharmacology databases for traditional chinese medicine: review and assessment. Front. Pharmacol. 10.
- Zhang, R.-Z., Yu, S.-J., Bai, H., Ning, K., 2017. TCM-Mesh: The database and analytical system for network pharmacology analysis for TCM preparations. Scientific Reports 7.
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2021.103384.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







